Volume 30, Number 11—November 2024
Research Letter
Outbreak of Listeriosis Likely Associated with Baker’s Yeast Products, Switzerland, 2022–2024
Cite This Article
Citation for Media
Abstract
We traced back a nationwide outbreak of human listeriosis in Switzerland to a persisting production line contamination of a factory producing baker’s yeast with Listeria monocytogenes serotype 1/2a sequence type 3141. We used whole-genome sequencing to match clinical isolates to isolates from product samples.
We report a prolonged and diffuse outbreak of listeriosis in Switzerland that involved 34 patients. The first case was reported in April 2022, and the outbreak peak occurred in 2023 (Figure 1). The last known case was in June 2024. A questionnaire-based outbreak investigation was initiated by the Swiss Federal Office of Public Health but did not identify a specific food exposure. The median age of the patients was 79 years (range 0–93 years); 18 (53%) were female and 16 (47%) male. The distribution of patients within Switzerland was wide (across different cantons) (Appendix Figure).
Of the 34 persons with documented cases, 7 died, and listeriosis was reported as the primary cause of death in all 7 cases according to the notification data. Of the 34 human Listeria monocytogenes isolates recovered, 28 were from blood, 2 from cerebrospinal fluid, and 1 from a swab specimen of a facial skin lesion; for 3 isolates, no information about the clinical sample was available.
We performed whole-genome sequencing (WGS) by using Illumina MiSeq next-generation sequencing technology (Illumina, https://www.illumina.com). We mapped sequencing reads against a multilocus sequence typing scheme based on 7 housekeeping genes and a 1,701-locus core genome multilocus sequence typing (cgMLST) scheme by using Ridom SeqSphere+ software version 9.0.2 (https://www.ridom.de/seqsphere) (1). We determined sequence types and cluster types upon submission to the L. monocytogenes cgMLST Ridom SeqSphere+ server (1).
We defined a cluster as a group of isolates with <10 different alleles between neighboring isolates (1). We assigned all 34 isolates to L. monocytogenes serotype 1/2a, sequence type 3141, cgMLST cluster type 18049; all isolates harbored fosX genes (coding for fosfomycin resistance) and vga(G) genes (coding for lincosamides and streptogramin A resistance). Furthermore, all isolates were genetically closely related (<2 allelic difference) to L. monocytogenes isolated from baker’s yeast (Saccharomyces cerevisiae) products from a commercial yeast factory and its production lines.
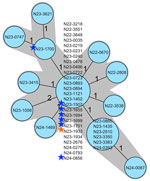
The baker’s yeast products were sold in retail stores and supplied the food industry throughout Switzerland. During 2022, the baker’s yeast manufacturer had determined the occurrence of L. monocytogenes in yeast product samples. Analysis had been conducted as part of the manufacturer’s self-control practices; however, in the absence of legal requirements for non–ready-to-eat (non-RTE) products, the findings were not reported. In July 2023, official random and risk-based food checks performed by a cantonal laboratory in Switzerland revealed the presence of L. monocytogenes (isolate N23-1507) in a yeast product. WGS analysis confirmed the presence of the outbreak strain (Figure 2). The finding prompted extensive environmental sampling on the production site of the manufacturer and retrospective WGS analysis of all available isolates.
During July 2023–August 2024, the manufacturer commissioned microbiological testing on different production steps, and we obtained product samples or swabs from the yeast cream, vats, pipe systems, drying filters, extruders, conveyor belts, and cutting machines. All sequenced L. monocytogenes isolated from the production line and the strains obtained from yeast products matched the outbreak strain (Figure 2). Within the production line, the laboratory identified a flow pipe connecting a starch vat to the yeast drying bed as the first point of contamination. Subsequently, the manufacturer started deep cleaning of all processing equipment at the part of the production line where the positivity was detected. An official product recall was not warranted because baker’s yeast is not classified as RTE food. However, only batches of the product that had been subjected to extensive controls and yielded <10 colony forming units of L. monocytogenes per gram were released for sale. This approach was supported by further investigations that showed no growth of L. monocytogenes during the shelf-life of the baker’s yeast (data not shown).
Products made with baker’s yeast normally undergo a heating step that would control the contamination. However, as evident from raw dough–associated illness outbreaks caused by Escherichia coli and Salmonella in Canada and the United States (2,3), contaminated dough represents a health hazard if consumed raw, and cross-contamination can occur through the handling of contaminated baker’s yeast or contaminated raw dough. That aspect is reflected by the fact that the outbreak strain also was detected in food items in several restaurants and institutional catering establishments in different cantons (data not shown).
Listeriosis is a potentially lethal infection, and the young, the elderly, pregnant women, and immunocompromised persons are at particular risk (4). Foods, mainly RTE foodstuffs that include meat, fish, dairy products, fruit, and vegetables, represent major vehicles for sporadic cases and outbreaks of listeriosis (5).
This outbreak, likely linked to baker’s yeast, highlights the lack of data on contamination of non-RTE products by foodborne pathogens, including L. monocytogenes, and the need for manufacturers of baker’s yeast to consider this risk in their production processes. Moreover, this outbreak should raise awareness that compliance with basic hygiene measures to prevent cross-contamination is particularly important when handling food that is not RTE.
Mr. Stephan is the director of the Institute for Food Safety and Hygiene at the University of Zurich. His research activities are focused on the epidemiology and characteristics of foodborne pathogens in the food chain.
Acknowledgments
We thank Nicole Cernela and Marc Stevens for the technical support with Illumina sequencing and bioinformatic analysis.
This work was partly supported by the Swiss Federal Office of Public Health’s Division Communicable Diseases and the Swiss Federal Office of Public Health’s Federal Food Safety and Veterinary Office.
The sequence data for clinical outbreak strain L. monocytogenes N23-0035 have been deposited in the National Center for Biotechnology Information Nucleotide database under BioProject no. PRJNA935533 and accession no. JBDQYW000000000.
References
- Ruppitsch W, Pietzka A, Prior K, Bletz S, Fernandez HL, Allerberger F, et al. Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes. J Clin Microbiol. 2015;53:2869–76. DOIPubMedGoogle Scholar
- Gill A, Carrillo C, Hadley M, Kenwell R, Chui L. Bacteriological analysis of wheat flour associated with an outbreak of Shiga toxin-producing Escherichia coli O121. Food Microbiol. 2019;82:474–81. DOIPubMedGoogle Scholar
- Rahman R, Scharff RL, Wu F. Foodborne disease outbreaks in flour and flour-based food products from microbial pathogens in the United States, and their health economic burden. Risk Anal. 2023;43:2519–26. DOIPubMedGoogle Scholar
- Allerberger F, Wagner M. Listeriosis: a resurgent foodborne infection. Clin Microbiol Infect. 2010;16:16–23. DOIPubMedGoogle Scholar
- Buchanan RL, Gorris LGM, Hayman MM, Jackson TC, Whiting RC. A review of Listeria monocytogenes: an update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control. 2017;75:1–13. DOIGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: September 25, 2024
Table of Contents – Volume 30, Number 11—November 2024
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Magdalena Nüesch-Inderbinen, Institute for Food Safety and Hygiene, Vetsuisse Faculty, University of Zurich, Winterthurerstrasse 272, 8057 Zurich, Switzerland
Top