Volume 30, Number 12—December 2024
Dispatch
Feline Panleukopenia Virus in a Marsican Brown Bear and Crested Porcupine, Italy, 2022–2023
Figure
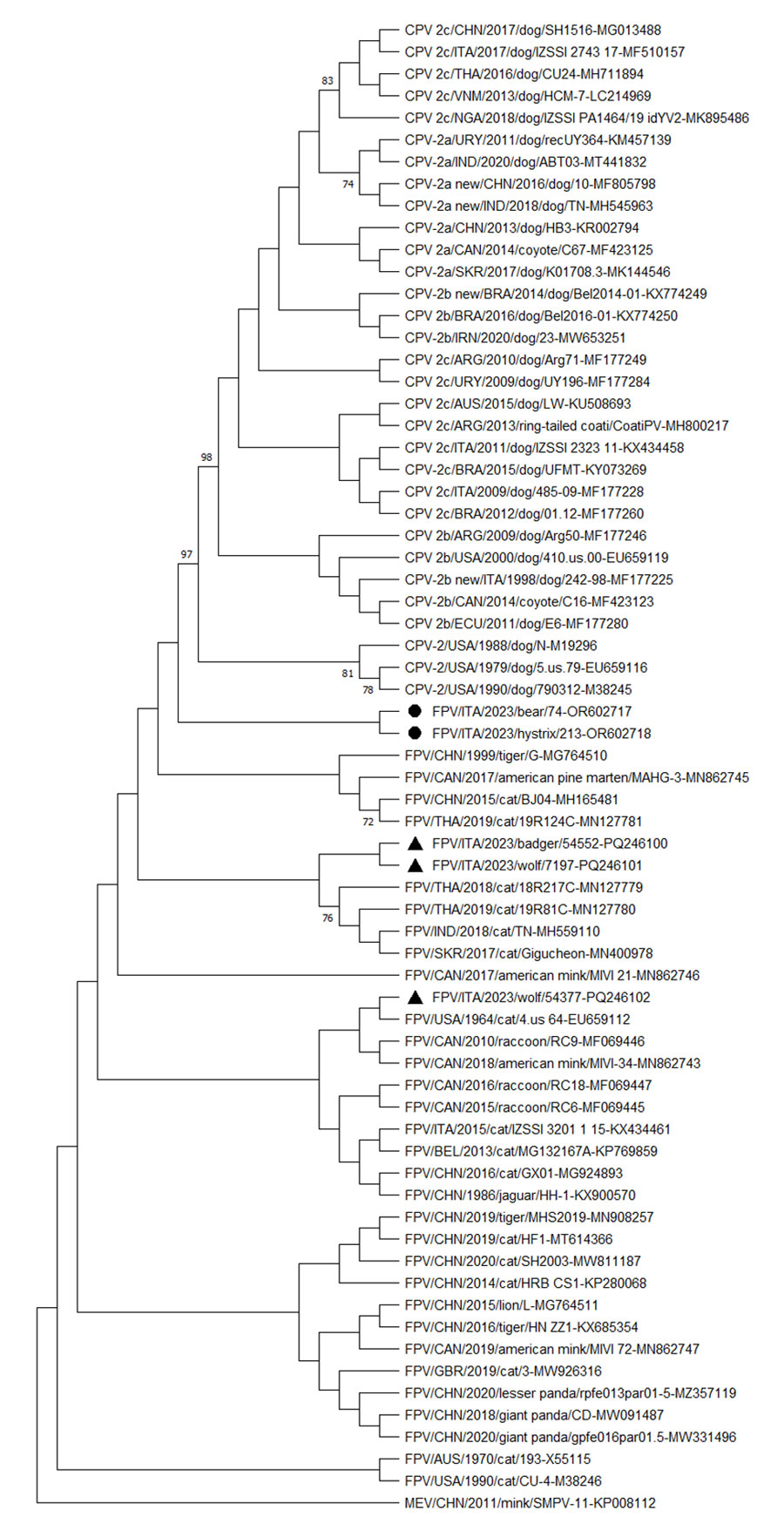
Figure. Neighbor-joining capsid protein 2 (VP2)–based phylogenetic tree of Protoparvovirus carnivoran 1 from study of feline panleukopenia virus (FPV) in wild animals, Abruzzo and Molise, Italy, January 2022–May 2023. The tree was elaborated by using a 584-aa long alignment of the VP2 sequences of the FPV strains generated in the study and the cognate sequences of Protoparvovirus carnivoran 1 strains retrieved from GenBank. The tree was constructed by using MEGA X version 10.0.5 software (https://www.megasoftware.net) and the maximum-likelihood method, the Jones-Taylor-Thornton substitution model, and bootstrapping up to 1,000 replicates. Bootstrap values >70% are shown. Black bullets indicate FPV strains from a Marsican brown bear (ΙΤΑ/2023/bear/74 (GenBank accession no. OR602717) and a crested porcupine ITA/2023/hystrix/213 (accession no. OR602718). The black triangles indicate FPV strains from other wildlife animals in the same study: ITA/2023/badger/54552 (accession no. PQ246100), ΙΤΑ/2023/wolf/7197 (accession no. PQ246101), and ΙΤΑ/2023/wolf/54377 (accession no. PQ246102). MEV (accession no. KP008112) was used as an outgroup. Scale bar indicates number of amino acid substitutions per site. CPV, canine parovirus; MEV, mink enteritis virus; FPV, feline panleukopenia virus.