Volume 30, Number 12—December 2024
Dispatch
Incursion of Novel Eurasian Low Pathogenicity Avian Influenza H5 Virus, Australia, 2023
Cite This Article
Citation for Media
Abstract
Australia is a sink for low pathogenicity avian influenza viruses, with isolated circulation occurring on the continent. We report the incursion of a Eurasian low pathogenicity avian influenza H5 virus into Australia. This report benefits surveillance and diagnostic work because of the risk and current absence of highly pathogenic avian influenza A(H5N1).
Australia is a sink for the diversity of avian influenza viruses, wherein viruses circulating in Eurasia and North America are occasionally introduced to the continent, followed by long-term, isolated circulation (1). Whereas a diversity of low pathogenicity avian influenza (LPAI) viruses exist, Oceania is the only continent where the goose/Guangdong lineage of highly pathogenic avian influenza (HPAI) A(H5N1) is absent (2). An HPAI incursion has never been detected in Oceania, although several HPAI outbreaks have occurred in poultry, all following evolution of endemic Australian lineage LPAI H7 viruses from the wild bird reservoir (3). Because of the risk to poultry and wild birds in Australia, a key aim of the National Avian Influenza in Wild Birds (NAIWB) surveillance program is the detection and characterization of circulating H5 and H7 viruses (4).
In 2023, through both the NAIWB targeted surveillance (4) and passive surveillance, we detected 17 LPAI H5 viruses by quantitative PCR (qPCR) and recovered 8 genomes. The genomes comprised 7 complete or near-complete genomes and 1 partial genome, which included hemagglutinin (HA), nucleoprotein, neuraminidase (NA), matrix, and nonstructural protein (NS) segments. We recovered viral HA fragments from 2 additional detections that identified lineage only (Table). We collected, screened, and sequenced samples as described in previous publications (1). We deposited the sequences into GenBank (GenBank accession nos. PP947529–52 and PP922943–79).
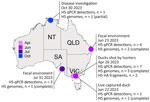
The HA segments of LPAI H5 viruses we sequenced were unrelated to LPAI H5 viruses previously characterized in Australia. The HA sequences had a PQRETR/GLF cleavage site, which contrasts with the PQKATR/GLF cleavage site found in all LPAI H5 viruses reported in Australia since 2005 (1). The detections we found all belonged to a single Eurasian-lineage LPAI H5 that was detected in 4 Australia jurisdictions over a 7-month period. We detected the first occurrence of the LPAI H5 in April 2023 (Figure 1) in Victoria from 3 hunter shot waterfowl: 1 Pacific black duck (Anas superciliosa), 1 gray teal (Anas gracilis), and 1 chestnut teal (Anas castanea). In June 2023, we detected another LPAI H5 occurrence in Victoria in a live-captured Pacific black duck and 2 LPAI H5 occurrences in Queensland from wild waterbird fecal environmental samples. We detected another LPAI H5 occurrence in July 2023 from wild waterbird fecal samples in South Australia. Finally, we detected 1 LPAI H5 virus in a wild duck in October 2023 through passive surveillance in the Northern Territory. Samples collected from live birds were done in accordance with Deakin University Animal Ethics (ethics approval no. B39-2019). Ethics approvals were not required for fecal environmental samples, samples collected from hunter-shot birds, or samples collected for disease investigations.
We conducted BLASTn analysis (https://blast.ncbi.nlm.nih.gov) by using the National Center for Biotechnology Information nucleotide database. We conducted phylogenetic analysis of the HA sequences and found the sequenced viruses were most closely related (<98%) to LPAI H5 viruses detected during 2019–2022 in Japan and South Korea (Figures 1, 2). Those countries contribute most influenza genomes recovered from the East Asian–Australasian databases. Alignments and .xml files for all trees are available at https://github.com/michellewille2/Eurasian-LPAI-H5-incursion-to-Australia. We investigated the potential temporal incursion window in Australia by using time-structured phylogenetic analysis, as outlined in previous publications (1). The most recent common ancestor of the 8 complete HA sequences was from April 2022 (95% highest posterior density [HPD] September 2021–October 2022), almost a year before the first detection, but with a large HPD (Figure 1). To examine H5 activity in the previous year, we queried all H5 detections for 2022 reported to the NAIWB surveillance program and found 12 LPAI H5 viruses were detected by qPCR in wild bird samples (New South Wales, n = 3; Tasmania, n = 1; Victoria, n = 5; Western Australia, n = 3) (5). Of the viruses sequenced, all HA sequences fell into the previously described Australian LPAI H5 lineage that has been circulating since 2007, although it was likely present since the 1990s (1). Hence, cryptic circulation of the novel Eurasian lineage LPAI H5 in Australia may have occurred in locations or in avian hosts that are not included in NAIWB surveillance and therefore were not detected in 2022. Alternatively, incursion might have only occurred in early 2023, but rather than a single viral introduction, multiple viruses with closely related HA sequences were separately introduced. Unfortunately, the scarcity of available HA reference sequences from 2022 and 2023 from Asia because of limitations in wild bird surveillance or sequence deposition into GenBank hinders understanding of introduction dynamics.
We investigated the reassortment patterns in the segments to gain additional insight. All other gene segments fall into viral lineages already present in Australia (Table; Appendix Figure, Tables 1, 2). Genome constellations were partially conserved, and all viruses shared the same polymerase basic (PB) 1, polymerase acidic, nucleoprotein, and matrix segment lineages, although within-clade segments were not always 100% identical (Appendix Figure). We detected 3 different PB1 lineages and 2 different NS lineages (both the A and B alleles). We obtained full genomes of 4 viruses from wild birds in Victoria, 3 from the same sampling event. Of the full genomes obtained, 2 had identical genome constellations, although A/gray teal/Victoria/23-01688-0045/2023 was a mixed infection with H5, H10, N3 and N7 segments recovered. Co-sampled A/chestnut Teal/Victoria/23–01686-0034/2023(H5N3) was different in both the PB1 and NS sequence compared with the other 2 viruses. A/Pacific black duck/Victoria/17363/2023(H5N3), which was detected 2 months after the initial detections in Victoria, had a similar genome constellation to the other sequences from Victoria, despite having an HA sequence sister to those from Queensland (detected June 2023), rather than those from Victoria. A/wild waterbird/South Australia/23–80999145–13/2023(H5N9) had a different PB1 and NA sequence, and A/radjah shelduck/Northern Territory/20231282–03/2023(H5N1) also had a different NA subtype. The 2 genomes from Queensland shared the same PB1 sequence as A/wild waterbird/South Australia/23-80999145-13/2023(H5N9) but had unique M segments. A reasonable explanation for those results is a single introduction followed by a reassortment event incorporating 7 Australian-lineage segments. As the virus spread across Australia, additional reassortment events contributed to the genome constellation diversity (i.e., diversity in the PB1, NA, and NS segments).
There are 3 implications of this viral incursion. First, the delayed detection highlights limitations in the national targeted surveillance program that, in combination with investigation of consequential wild bird illness and death events, is necessary to rapidly detect and respond to an incursion of goose/Guangdong HPAI H5N1 into Australia. Second, while the KATR cleavage site found in Australian-lineage viruses has not yet been associated with evolution of LPAI to HPAI, the RETR cleavage site in the novel Eurasian lineage incursion has been identified as a precursor to HPAI H5 viruses (6). This incursion may increase the risk for evolution of HPAI H5 in Australia. Finally, as part of the surveillance and diagnostic processes, H5 qPCRs are designed to rapidly differentiate LPAI H5 of Australian lineage from Eurasian lineage LPAI and HPAI. The presence of a novel LPAI lineage requires a diagnostic update. Understanding viral incursions is a global concern, as demonstrated by the numerous incursions and intercontinental spread of HPAI H5N1 clade 2.3.4.4b. Determining the patterns of viral incursion and spread and identifying the hosts involved is necessary for effective disease risk management and communication that supports disease preparedness and response.
Dr. Wille is a senior research fellow at the Centre for Pathogen Genomics, University of Melbourne. Her interests include viruses of birds, ecology and evolution of avian influenza, and ecology of bird viral communities.
Acknowledgments
We thank the jurisdictions, programs, and individuals responsible for sharing baseline H5 data for 2022 and 2023.
The National Avian Influenza in Wild Birds Program and the Australian Centre for Disease Preparedness are funded by Department of Agriculture Fisheries and Forestry. The World Health Organization Collaborating Centre for Reference and Research on Influenza is supported by the Department for Health and Aged Care.
References
- Wille M, Grillo V, Ban de Gouvea Pedroso S, Burgess GW, Crawley A, Dickason C, et al. Australia as a global sink for the genetic diversity of avian influenza A virus. PLoS Pathog. 2022;18:
e1010150 . DOIPubMedGoogle Scholar - Wille M, Atkinson R, Barr IG, Burgoyne C, Bond AL, Boyle D, et al. Long-distance avian migrants fail to bring 2.3.4.4b HPAI H5N1 into Australia for a second year in a row. Influenza Other Respir Viruses. 2024;18:
e13281 . DOIPubMedGoogle Scholar - Scott A, Hernandez-Jover M, Groves P, Toribio JA. An overview of avian influenza in the context of the Australian commercial poultry industry. One Health. 2020;10:
100139 . DOIPubMedGoogle Scholar - Grillo VL, Arzey KE, Hansbro PM, Hurt AC, Warner S, Bergfeld J, et al. Avian influenza in Australia: a summary of 5 years of wild bird surveillance. Aust Vet J. 2015;93:387–93. DOIPubMedGoogle Scholar
- National Avian Influenza Wild Bird Program. Wild bird news, issue 9. 2023 [cited 2024 Sep 12]. https://wildlifehealthaustralia.com.au/Resource-Centre/Surveillance-Reports
- Luczo JM, Stambas J, Durr PA, Michalski WP, Bingham J. Molecular pathogenesis of H5 highly pathogenic avian influenza: the role of the haemagglutinin cleavage site motif. Rev Med Virol. 2015;25:406–30. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: November 01, 2024
Table of Contents – Volume 30, Number 12—December 2024
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Michelle Wille, University of Melbourne,792 Elizabeth St, Melbourne, VIC 3000, Australia
Top