Volume 30, Supplement—October 2024
SUPPLEMENT ISSUE
Articles
Comprehensive Surveillance of Severe Fever with Thrombocytopenia Syndrome Virus in Patients with Acute Febrile Illness, Wild Rodents, and Trombiculid Larval Mites, Thailand
Abstract
Infection with severe fever with thrombocytopenia syndrome (Bandavirus dabieense) virus poses a substantial public health threat because of its high mortality rates and severe complications. The virus is prevalent in Asia, although data from Thailand are scarce. Our study confirmed the virus in 1.6% of acute febrile illness patients and specific antibodies in 3% of archived samples since 2015 in Thailand. Nationwide zoonotic surveillance identified the virus in 8 rodent species and 4 chigger genera. Our findings underscore the importance of raising awareness among healthcare providers and the general public about the symptoms, risks, and prevention strategies associated with severe fever with thrombocytopenia syndrome virus infection. Ongoing surveillance of the virus in human and animal populations is essential for monitoring its prevalence, distribution, and potential for emergence.
Severe fever with thrombocytopenia syndrome virus (SFTSV) is a virulent virus with a triple-segmented, negative-sense, single-stranded RNA genome. Taxonomically Bandavirus dabieense, the virus belongs to the genus Bandavirus, family Phenuiviridae. SFTSV poses a substantial public health challenge because of the lack of a vaccine or effective therapies and high mortality rates in previously healthy persons (1–3). First discovered in China in 2009 (4), SFTSV has since been reported in China, South Korea, and Japan and more recently in Vietnam, Myanmar, Pakistan, Taiwan, and Thailand (5–12). The virus is classified into genotypes A–F, each having distinct geographic variations in virulence and pathogenicity (13–15).
SFTSV is predominantly transmitted through tick bites, specifically by the Asian longhorned tick Haemaphysalis longicornis, known for its wide host range, vector competency for various pathogens, and extensive geographic distribution (16). Additional competent vectors include Haemaphysalis flava (17), Ixodes sinensis (18), and >8 other implicated tick species (19,20). Tick-bite prevention is considered the primary means of preventing SFTSV infection. Evidence also implicates mites in SFTSV transmission, particularly those in the family Trombiculidae, including Leptotrombidium scutellare and Leptotrombidium deliense, and family Laelapidae, including Laelaps echidninus (21,22).
Human-to-human transmission of SFTSV occurs through direct contact with infected blood or bodily fluids (23). Animal-to-human transmission is occasionally reported though contact with ill animals (12,24). The role of wild and domesticated animals has garnered considerable interest because of their potential involvement as pathogen reservoirs. In addition, the presence of viral RNA or specific antibodies has been confirmed in 10 domestic (20,25–27) and >10 wild animal species (28–30). This broad host and vector involvement underlines the complex epidemiology of SFTSV, posing major challenges in developing targeted public health strategies and mitigating the effect of this virus. Among vertebrate reservoirs of concern, rodents receive considerable attention because of their close proximity to humans.
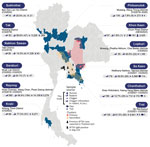
Although SFTSV in Thailand was documented in 2020 (12), evidence suggests that the presence of SFTSV dates back to 2019 (31). Subsequent analysis of patients clinically suspected of having viral infection confirmed the presence of SFTSV genome segments belonging to genotype B (32). One study in Thailand investigated dogs as amplifying hosts, and the nucleotide sequence of SFTSV found in 1 dog appears closely related to genotype B or J3 (33). Although evidence from human and animal studies indicates the presence of SFTSV in multiple provinces of Thailand, primarily in Bangkok and its neighboring regions (Figure 1), the understanding of its distribution and potential hotspots remains incomplete. Comprehensive surveillance, particularly from regions with extensive farming or agricultural activities, where these habitats could serve as zoogeographic transmission points and disease hotspots, is imperative to guide effective prevention and control strategies.
To better understand the epidemiologic effect of SFTSV in Thailand, our study evaluated the prevalence of SFTSV RNA and specific antibodies in serum samples from 2,425 patients with undifferentiated acute febrile illness (AFI). The patients were admitted to Chum Phae Hospital in Khon Kaen Province, northeastern Thailand, during 2015–2021. We performed RNA detection by using quantitative reverse transcription PCR (qRT-PCR) targeting the partial nonstructural (NS) protein–encoding gene of the small segment (Appendix). We subsequently screened all SFTSV RNA-positive samples for other vectorborne pathogens endemic in Southeast Asia, including dengue, chikungunya, and Zika virus, by using the ZDC Multiplex RT-PCR Assay Kit (Bio-Rad, https://www.bio-rad.com) and bacterial pathogens, including Rickettsia and Orientia spp., by following a previously described protocol (34). We verified all qRT-PCR products for the expected size and subjected them to nucleotide sequencing by using the barcode-tagged sequencing method (Bionics, https://www.bionicsro.co.kr). We conducted phylogenetic analysis by using MEGA version 11 (https://www.megasoftware.net). The resulting nucleotide sequences of SFTSV amplified from the samples have been deposited into the GenBank database (accession nos. PP782658–61).
We detected SFTSV RNA in 38 of 2,425 AFI patients, resulting in a positivity rate of 1.6%. The median age of SFTSV RNA–positive patients was 47.2 years (15.9–86.7 years) (first–third interquartile range [IQR1–3] 29–64 years). A total of 36.8% of the patients had agriculture-related occupations; male:female ratio was 1.4:1.0 (Table 1). The SFTSV RNA–positive cases did not exhibit a discernible seasonal pattern. We observed no co-positivity between SFTSV and other pathogens. SFTSV RNA–positive patients often visited the hospital with fever (38.0°C [range 36.1°C–40.3°C]); 20.5% experienced headaches, and 12.8% reported dizziness. We observed thrombocytopenia and variations in hemoconcentration in 3 cases (Appendix Figure 1). Of note, all patients recovered without severe complications. We successfully obtained only 2 nucleotide sequences from SFTSV RNA–positive patients, both of which demonstrated a close genetic relationship to genotype D. The strains shared 99.1% nucleotide similarity to strain LN2012–41 (GenBank accession no. KF887433) previously identified in a patient in China in 2012 (Figure 2).
We used an indirect ELISA specific for nucleoprotein (NP) of SFTSV to detect SFTSV IgM and SFTSV IgG in all 2,425 serum samples of AFI patients, according to manufacturer instructions (Bore Da Biotech, http://boreda.com). The average optical density at 450 nm (OD450) of the positive controls provided by the kit was 0.233 for SFTSV IgM and 0.172 for SFTSV IgG. We considered samples with OD450 values exceeding those cutoffs to be seropositive (Appendix Figure 2). Serologic analysis revealed that 16 patients (0.7%) were seropositive for SFTSV IgM and 54 patients (2.2%) were seropositive for SFTSV IgG; 3 patients (0.1%) had detectable levels of both antibodies (Appendix Figure 2). Those findings resulted in an overall seropositive rate of 3% among the study population (Table 1). The median age of SFTSV IgM–seropositive patients was 63.8 years (IQR1–3 20.6–68.1 years) and for SFTSV IgG–seropositive patients was 14 years (IQR1–3 8.4–51.5 years). We observed no co-positive results between ELISA and qRT-PCR in the tested samples.
We evaluated the practicality of the paper-based lateral flow immunochromatography rapid test (SFTSV RDT; Bore Da Biotech) for potential use in prescreening of the viral NP at the point-of-care. Of the 38 SFTSV RNA–positive serum samples, 33 samples had sufficient quantities and were included in the analysis. The SFTSV RDT accurately detected the viral NP when the samples had a qRT-PCR cycle threshold of <37 or an average of 2.72 × 104 copies/mL serum observed in our study, demonstrating a positive concordance rate of 89.5% between both assays. Control tests with serum from healthy donors and patient serum samples previously confirmed positive for dengue virus 1–4, chikungunya virus, Rickettsia typhi, or Orientia tsutsugamushi showed no cross-reactivity. The overall agreement between SFTSV RDT and qRT-PCR was substantial, having a κ value of 0.732, which validated its effectiveness (Appendix Table 3).
To investigate the role of rodents and chiggers in SFTSV transmission, we analyzed a total of 2,052 tissue samples from 1,019 wild rodents, representing 15 species across 7 genera and 4 families, by using qRT-PCR targeting the NS gene. We collected the samples during 2019–2023 as part of the rodentborne and ectoparasiteborne disease risk assessment program in Thailand (Appendix). Eleven rodents from 8 species were positive for SFTSV RNA, indicating an overall positivity rate of 1.1%. We collected the RNA-positive samples during 2019–2023 (Appendix Table 1) in Nakhon Sawan, Chanthaburi, Sa Kaeo, and Khon Kaen Provinces, and the positive rodent species included Mus caroli (Ryukyu mouse), Menetes berdmorei (Berdmore’s squirrel), Rattus norvegicus (Norway rat), Berylmys berdmorei (small white-toothed rat), Rattus exulans (Polynesian rat), Bandicota indica (greater bandicoot rat), Bandicota savilei (Savile’s bandicoot rat), and Rattus tanezumi complex (Asian house rat) (Table 2). Of the 1,019 rodents analyzed, we observed the highest SFTSV RNA prevalence in lungs (0.4% [4 rodents]) and spleen (0.4% [4 rodents]), followed by kidneys (0.3% [3 rodents]). Phylogenetic analysis of the partial sequence of the NS gene obtained from 2 rodents indicated a close relationship to genotype D, sharing 97.3% and 99.1% nucleotide similarity to the strain LN2012–41 from China and being nearly identical to SFTSV sequences obtained from AFI patients (Figure 2). The high genetic similarity observed across different locations and periods suggests a potential widespread distribution of SFTSV in Thailand. However, this observation might also be influenced by the limitation of using only 124-bp nucleotide sequences for phylogenetic analysis. To ensure the validity of our findings and to rule out the possibility of cross-contamination, we performed nucleic acid extraction of samples and qRT-PCR in 2 separate laboratories by using different stocks of samples and reagents for confirmation. In addition, we prepared the positive control for the assay by using virus culture stock from an SFTSV RNA–positive case in South Korea that displayed a genetic distance from our positive samples.
From the analysis of 573 individual chiggers retrieved from 155 wild rodents, we detected SFTSV RNA in 8 chiggers (1.4%), which had an average SFTSV RNA level of 2.40 × 104 copies/chigger (range 5.80 × 103–70 × 104 copies/chigger) (Appendix Table 2). We retrieved the SFTSV RNA–positive chiggers from 6 rodents from Sa Kaeo, Chanthaburi, Lopburi, Rayong, and Trat provinces. Phylogenetic analysis of cytochrome oxidase subunit I gene sequences confirmed the correct genus assignment for SFTSV RNA–positive chiggers and revealed close relatedness to Gahrliepia (walchia) (4 samples), Blankaartia acuscutellaris (1 sample), and Schoengastia kanhaensis mites (1 sample) (Appendix Figure 3). Two SFTSV RNA–positive chiggers from the genus Leptotrombidium had insufficient sample quantities for further analysis. In addition, 4 of 8 SFTSV RNA–positive chiggers showed co-positivity for unidentified Rickettsia, but none tested positive for O. tsutsugamushi.
Our study confirmed the presence of SFTSV RNA in AFI patients, wild rodents, and chiggers across multiple locations in Thailand and identified a notable seroprevalence of SFTSV-specific antibodies among AFI patients, highlighting a substantial yet underrecognized prevalence of SFTSV. The analysis of samples spanning multiple years, including the detection of SFTSV RNA in AFI patients in 2015, suggests that SFTSV has been circulating in Thailand since 2015, which predates the SFTSV detections reported in other Southeast Asia countries, including Vietnam in 2017 (35), Thailand in 2019 (12,31) and Myanmar during 2018–2019 (11). Our findings provide additional evidence of the existence of SFTSV genotype D, indicating that >2 genotypes have been identified recently in Thailand. Positive cases in patients and animals have been identified across several provinces in multiple regions, including Chachoengsao, Samutprakan, and Chonburi Provinces in the central region; Nakhon Sawan Province in the northern region; Chanthaburi, Sa Kaeo, Prachinburi, Rayong, and Chonburi Provinces in the eastern region; and Khon Kaen Province in the northeastern region (Figure 1) (31–33). This extensive distribution of the virus signifies a widespread and longstanding effect, necessitating ongoing surveillance and enhanced diagnostic measures to fully comprehend the disease ecology and transmission dynamics and to manage public health interventions effectively.
We observed that the prevalence of SFTSV RNA in wild rodents in our study was similar to the rate reported in China, where 0.7% of rodents tested positive. The species in China included Apodemus agrarius (striped field mouse), Crocidura lasiura (Ussuri white-toothed shrew), R. norvegicus (brown rat), and Mus musculus (house mouse) (22). A subsequent study from the same authors reported a higher prevalence of 32.3% and positive species including A. agrarius mice, Tscherskia triton (greater long-tailed hamster), and M. musculus mice (14). The primary difference between our study and previous studies lies in the surveillance sites and periods. In our study, rodents were captured from semirural areas near communities and were primarily collected during the dry season in Thailand within a 2–3-month timeframe at selected sites. During this period, agricultural activities in some locations are reduced, which could potentially limit rodent abundance and their exposure to a broader range of ectoparasites, including ticks. To more accurately determine whether rodents serve as reservoirs of SFTSV and better understand the complex dynamics of SFTSV transmission, comprehensive surveillance studies involving collection of multiple rodent species across diverse geographic locations, seasons, and rodent species, coupled with serologic analysis, are crucial. Incorporating factors such as habitat diversity, agricultural practices, and climatic conditions into future research will also contribute to a comprehensive understanding of the ecologic niche of SFTSV.
Of note, we detected SFTSV RNA in chiggers infesting rodents that tested negative for the virus. This unexpected finding suggests a potential lack of direct rodent-to-chigger transmission. Given that chiggers feed on liquefied host skin tissue rather than blood, accidental acquisition of the virus during feeding appears less likely. Experimental studies supporting this notion demonstrated that chiggers feeding on O. tsutsugamushi–infected hosts failed to transmit the pathogen to their offspring (36). Alternatively, chiggers may acquire the virus through co-feeding with infected conspecifics and subsequently transmit the virus transovarially to their offspring, a phenomenon that has been successfully demonstrated in establishing new lines of O. tsutsugamushi–infected chiggers in previous research at our institute (37). Furthermore, reported co-positivity for SFTSV and O. tsutsugamushi in patients from South Korea and Myanmar (11,38,39) suggests a potential epidemiologic intersection between these pathogens, possibly enabled by shared vector species or overlapping habitats (40). The co-concurrence of these infections complicates diagnosis and underscores the importance of integrated surveillance systems to monitor these and potentially other co-circulating pathogens.
In conclusion, although the role of chiggers in SFTSV transmission remains unclear, the widespread distribution and abundance of chiggers, especially in the Asia–Pacific region, leads to frequent exposure to chiggerborne pathogens. Our findings suggest that the epidemiology of SFTSV may be more complex than previously understood, involving several potential vectors, reservoir hosts, and interactions with other pathogens. This apparent complexity underscores the need for comprehensive surveillance and research to better understand and mitigate the risks associated with this emerging infectious disease.
Dr. Linsuwanon is chief of the Arthropod-Borne and Zoonotic Diseases section at the Department of Entomology, Walter Reed Army Institute of Research–Armed Forces Research Institute of Medical Sciences. Her primary research interest focuses on operational risk assessment for ectoparasiteborne and rodentborne diseases.
Acknowledgments
We extend our sincere gratitude to the medical personnel and physicians at Chum Phae Hospital in Khon Kaen Province and the Center of Excellence in Clinical Virology at Chulalongkorn University in Bangkok for their invaluable contributions in specimen collection, processing, and data sharing. We are grateful for the support from Wuttikon Rodhvamtook, Royal Thai Army–Armed Forces Research Institute of Medical Sciences in the surveillance in military installation in Lopburi Province; Taweesak Monkanna; and Opas Thachin; as well as local hunters and property owners for their assistance in performing field-rodent sample collection.
This study was an integral part of the operational entomological risk assessment conducted to support military training exercise in the Indo-Pacific region supported by the US Department of Defense Armed Forces Health Surveillance Division Global Emerging Infections Surveillance Program (proposal identification nos. P0085_21_AF, P0092_22_AF, P0035_23_AF, and P0035_24_AF).
Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation or publication. The opinions or assertions contained herein are the private views of the author, and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense. Research was conducted under an approved animal use protocol in an Association for Assessment and Accreditation of Laboratory Animal Care–accredited facility in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals and adheres to principles stated in the National Research Councils Guide for the Care and Use of Laboratory Animals (41). The investigators have adhered to the policies for protection of human subjects as prescribed in Army Regulation 70–25 (42).
References
- Wang X, Ren X, Ge Z, Cui S, Wang L, Chen Z, et al. Clinical manifestations of death with severe fever and thrombocytopenia syndrome: A meta-analysis and systematic review. J Med Virol. 2021;93:3960–8. DOIPubMedGoogle Scholar
- Choi SJ, Park SW, Bae IG, Kim SH, Ryu SY, Kim HA, et al.; for Korea SFTS Clinical Network. for Korea SFTS Clinical Network. Severe fever with thrombocytopenia syndrome in South Korea, 2013–2015. PLoS Negl Trop Dis. 2016;10:
e0005264 . DOIPubMedGoogle Scholar - Kobayashi Y, Kato H, Yamagishi T, Shimada T, Matsui T, Yoshikawa T, et al.; SFTS Epidemiological Research Group Japan. Severe fever with thrombocytopenia syndrome, Japan, 2013–2017. Emerg Infect Dis. 2020;26:692–9. DOIPubMedGoogle Scholar
- Cai L, Zhang H, Gao LD, Hu SX, Xie LY, Zhan ZF, et al. Identification of the first case of SFTSV infection in the Hunan Province of China and epidemiological surveillance in the locality. Ticks Tick Borne Dis. 2019;10:454–61. DOIPubMedGoogle Scholar
- Kurihara S, Satoh A, Yu F, Hayasaka D, Shimojima M, Tashiro M, et al. The world first two cases of severe fever with thrombocytopenia syndrome: An epidemiological study in Nagasaki, Japan. J Infect Chemother. 2016;22:461–5. DOIPubMedGoogle Scholar
- Park SW, Ryou J, Choi WY, Han MG, Lee WJ. Epidemiological and clinical features of severe fever with thrombocytopenia syndrome during an outbreak in South Korea, 2013–2015. Am J Trop Med Hyg. 2016;95:1358–61. DOIPubMedGoogle Scholar
- Tran XC, Yun Y, Van An L, Kim SH, Thao NTP, Man PKC, et al. Endemic severe fever with thrombocytopenia syndrome, Vietnam. Emerg Infect Dis. 2019;25:1029–31. DOIPubMedGoogle Scholar
- Takahashi T, Maeda K, Suzuki T, Ishido A, Shigeoka T, Tominaga T, et al. The first identification and retrospective study of Severe Fever with Thrombocytopenia Syndrome in Japan. J Infect Dis. 2014;209:816–27. DOIPubMedGoogle Scholar
- Chen S, Saqib M, Khan HS, Bai Y, Ashfaq UA, Mansoor MK, et al. Risk of infection with arboviruses in a healthy population in Pakistan based on seroprevalence. Virol Sin. 2024;39:369–77. DOIPubMedGoogle Scholar
- Lin TL, Ou SC, Maeda K, Shimoda H, Chan JP, Tu WC, et al. The first discovery of severe fever with thrombocytopenia syndrome virus in Taiwan. Emerg Microbes Infect. 2020;9:148–51. DOIPubMedGoogle Scholar
- Win AM, Nguyen YTH, Kim Y, Ha NY, Kang JG, Kim H, et al. Genotypic heterogeneity of Orientia tsutsugamushi in scrub typhus patients and thrombocytopenia syndrome co-infection, Myanmar. Emerg Infect Dis. 2020;26:1878–81. DOIPubMedGoogle Scholar
- Ongkittikul SW, Watanawong R, Rompho R. Severe fever with thrombocytopenia syndrome virus: the first case report in Thailand. Bangk Med J. 2020;16:204–6. DOIGoogle Scholar
- Yoshikawa T, Shimojima M, Fukushi S, Tani H, Fukuma A, Taniguchi S, et al. Phylogenetic and geographic relationships of severe fever with thrombocytopenia syndrome virus in China, South Korea, and Japan. J Infect Dis. 2015;212:889–98. DOIPubMedGoogle Scholar
- Lam TT, Liu W, Bowden TA, Cui N, Zhuang L, Liu K, et al. Evolutionary and molecular analysis of the emergent severe fever with thrombocytopenia syndrome virus. Epidemics. 2013;5:1–10. DOIPubMedGoogle Scholar
- Dai ZN, Peng XF, Li JC, Zhao J, Wu YX, Yang X, et al. Effect of genomic variations in severe fever with thrombocytopenia syndrome virus on the disease lethality. Emerg Microbes Infect. 2022;11:1672–82. DOIPubMedGoogle Scholar
- Luo LMZL, Zhao L, Wen HL, Zhang ZT, Liu JW, Fang LZ, et al. Haemaphysalis longicornis ticks as reservoir and vector of severe fever with thrombocytopenia syndrome virus in China. Emerg Infect Dis. 2015;21:1770–6. DOIPubMedGoogle Scholar
- Fang LZXX, Xiao X, Lei SC, Liu JW, Yu XJ. Haemaphysalis flava ticks as a competent vector of severe fever with thrombocytopenia syndrome virus. Ticks Tick Borne Dis. 2023;14:
102100 . DOIPubMedGoogle Scholar - Hu YY, Zhuang L, Liu K, Sun Y, Dai K, Zhang XA, et al. Role of three tick species in the maintenance and transmission of Severe Fever with Thrombocytopenia Syndrome Virus. PLoS Negl Trop Dis. 2020;14:
e0008368 . DOIPubMedGoogle Scholar - Luo LM, Zhao L, Wen HL, Zhang ZT, Liu JW, Fang LZ, et al. Haemaphysalis longicornis ticks as reservoir and vector of severe fever with thrombocytopenia syndrome virus in China. Emerg Infect Dis. 2015;21:1770–6. DOIPubMedGoogle Scholar
- Wang JN, Li TQ, Liu QM, Wu YY, Luo MY, Gong ZY. Vectors, hosts, and the possible risk factors associated with severe fever with thrombocytopenia syndrome. Can J Infect Dis Med Microbiol. 2021;2021:
8518189 . DOIPubMedGoogle Scholar - Gu XL, Su WQ, Zhou CM, Fang LZ, Zhu K, Ma DQ, et al. SFTSV infection in rodents and their ectoparasitic chiggers. PLoS Negl Trop Dis. 2022;16:
e0010698 . DOIPubMedGoogle Scholar - Wang QK, Ge HM, Hu JL, Zhang ZY, Wang YP, Jiao YJ, et al. Surveillance of vectors and host animals of severe fever with thrombocytopenia syndrome virus in Donghai, China in 2010–2011 [in Chinese]. Zhongguo Meijie Shengwuxue Ji Kongzhi Zazhi. 2013;4:313–6.
- Fang X, Hu J, Peng Z, Dai Q, Liu W, Liang S, et al. Epidemiological and clinical characteristics of severe fever with thrombocytopenia syndrome bunyavirus human-to-human transmission. PLoS Negl Trop Dis. 2021;15:
e0009037 . DOIPubMedGoogle Scholar - Yamanaka A, Kirino Y, Fujimoto S, Ueda N, Himeji D, Miura M, et al. Direct transmission of severe fever with thrombocytopenia syndrome virus from domestic cat to veterinary personnel. Emerg Infect Dis. 2020;26:2994–8. DOIPubMedGoogle Scholar
- Niu G, Li J, Liang M, Jiang X, Jiang M, Yin H, et al. Severe fever with thrombocytopenia syndrome virus among domesticated animals, China. Emerg Infect Dis. 2013;19:756–63. DOIPubMedGoogle Scholar
- Chen C, Li P, Li KF, Wang HL, Dai YX, Cheng X, et al. Animals as amplification hosts in the spread of severe fever with thrombocytopenia syndrome virus: A systematic review and meta-analysis. Int J Infect Dis. 2019;79:77–84. DOIPubMedGoogle Scholar
- Kuan CY, Lin TL, Ou SC, Chuang ST, Chan JP, Maeda K, et al. The first nationwide surveillance of severe fever with thrombocytopenia syndrome in ruminants and wildlife in Taiwan. Viruses. 2023;15:441. DOIPubMedGoogle Scholar
- Yun Y, Heo ST, Kim G, Hewson R, Kim H, Park D, et al. Phylogenetic analysis of severe fever with thrombocytopenia syndrome virus in South Korea and migratory bird routes between China, South Korea, and Japan. Am J Trop Med Hyg. 2015;93:468–74. DOIPubMedGoogle Scholar
- Oh SS, Chae JB, Kang JG, Kim HC, Chong ST, Shin JH, et al. Detection of severe fever with thrombocytopenia syndrome virus from wild animals and Ixodidae ticks in the Republic of Korea. Vector Borne Zoonotic Dis. 2016;16:408–14. DOIPubMedGoogle Scholar
- Kaneko C, Mekata H, Umeki K, Sudaryatma PE, Irie T, Yamada K, et al. Seroprevalence of severe fever with thrombocytopenia syndrome virus in medium-sized wild mammals in Miyazaki, Japan. Ticks Tick Borne Dis. 2023;14:
102115 . DOIPubMedGoogle Scholar - Rattanakomol P, Khongwichit S, Linsuwanon P, Lee KH, Vongpunsawad S, Poovorawan Y. Severe fever with thrombocytopenia syndrome virus infection, Thailand, 2019–2020. Emerg Infect Dis. 2022;28:2572–4. DOIPubMedGoogle Scholar
- Rattanakomol P, Khongwichit S, Chuchaona W, Vongpunsawad S, Poovorawan Y. Severe fever with thrombocytopenia syndrome virus genotype B in Thailand. Arch Virol. 2023;168:271. DOIPubMedGoogle Scholar
- Ishijima K, Phichitraslip T, Naimon N, Ploypichai P, Kriebkajon B, Chinarak T, et al. High seroprevalence of severe fever with thrombocytopenia syndrome virus infection among the dog population in Thailand. Viruses. 2023;15:2403. DOIPubMedGoogle Scholar
- Jitvaropas R, Sawaswong V, Poovorawan Y, Auysawasdi N, Vuthitanachot V, Wongwairot S, et al. Identification of bacteria and viruses associated with patients with acute febrile illness in Khon Kaen Province, Thailand. Viruses. 2024;16:630. DOIPubMedGoogle Scholar
- Tran XC, Kim SH, Lee JE, Kim SH, Kang SY, Binh ND, et al. Serological evidence of severe fever with thrombocytopenia syndrome virus and IgM positivity were identified in healthy residents in Vietnam. Viruses. 2022;14:2280. DOIPubMedGoogle Scholar
- Traub R, Wisseman CL Jr, Jones MR, O’Keefe JJ. The acquisition of Rickettsia tsutsugamushi by chiggers (trombiculid mites) during the feeding process. Ann N Y Acad Sci. 1975;266:91–114. DOIPubMedGoogle Scholar
- Frances SP, Watcharapichat P, Phulsuksombati D, Tanskul P. Transmission of Orientia tsutsugamushi, the aetiological agent for scrub typhus, to co-feeding mites. Parasitology. 2000;120:601–7. DOIPubMedGoogle Scholar
- Chatterjee S, Kim CM, Kim DM, Seo JW, Kim DY, Yun NR, et al. Coinfection with severe fever with thrombocytopenia syndrome and scrub typhus in Korea. Open Forum Infect Dis. 2023;10:
ofad377 . DOIPubMedGoogle Scholar - Thi Hai Yen N, Kim C, Jeong S, Jeon K, Choi H, Ro HJ, et al. Severe fever with thrombocytopenia syndrome virus infection or mixed infection with scrub typhus in South Korea in 2000–2003. Am J Trop Med Hyg. 2019;101:1096–9. DOIPubMedGoogle Scholar
- Miao D, Liu MJ, Wang YX, Ren X, Lu QB, Zhao GP, et al. Epidemiology and ecology of severe fever with thrombocytopenia syndrome in China, 2010‒2018. Clin Infect Dis. 2021;73:e3851–8. DOIPubMedGoogle Scholar
- National Research Council. Guide for the care and use of laboratory animals. 8th edition. Washington: National Academies Press; 2011 [cited 2024 May 31]. https://grants.nih.gov/grants/olaw/guide-for-the-care-and-use-of-laboratory-animals.pdf
- US Department of the Army. Army regulation 70–25. Research and development: use of volunteers as subjects of research. Washington: Department of the Army; 1990 [cited 2024 May 31]. https://armypubs.army.mil/epubs/DR_pubs/DR_a/pdf/web/r70_25.pdf
Figures
Tables
Cite This ArticleOriginal Publication Date: November 11, 2024
Table of Contents – Volume 30, Supplement—October 2024
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Piyada Linsuwanon, Walter Reed Army Institute of Research–Armed Forces Research Institute of Medical Sciences, Department of Entomology, 315/6 Rajavithi Rd, Thung Phayathai, Patumwan, Bangkok 10400, Thailand
Top