Volume 30, Supplement—October 2024
SUPPLEMENT ISSUE
Articles
Using SARS-CoV-2 Sequencing Data to Identify Reinfection Cases in the Global Emerging Infections Surveillance Program, United States
Figure 5
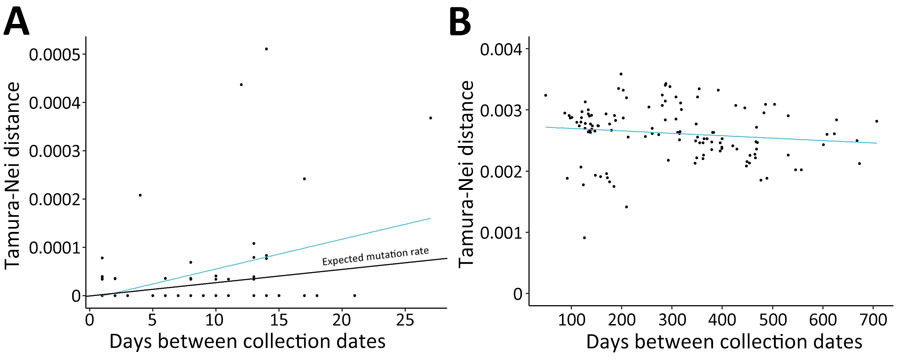
Figure 5. SARS-CoV-2 nucleotide changes in study using sequencing data to identify reinfection cases in Department of Defense Global Respiratory Pathogen Surveillance Program, United States. Tamura-Nei p-distances were determined relative to the number of days between specimen collection dates for continuing infections (A) and reinfections (B). A) Number of nucleotide substitutions correlated with the amount of time between specimen collections in patients who had continuing infections (p = 0.0021). Expected SARS-CoV-2 mutation rate was 1 single nucleotide variant per 2 weeks. B) No relationship was observed between number of nucleotide substitutions and time in reinfection cases (p = 0.137).
Page created: October 08, 2024
Page updated: November 11, 2024
Page reviewed: November 11, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.