Volume 30, Supplement—October 2024
SUPPLEMENT ISSUE
Articles
Azithromycin Resistance Patterns in Escherichia coli and Shigella before and after COVID-19, Kenya
Figure 2
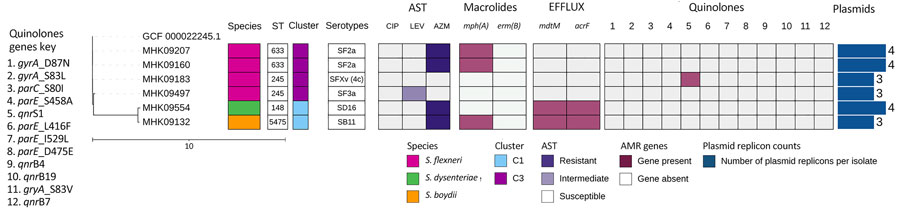
Figure 2. Phylogenetic tree and corresponding heatmap of 6 Shigella spp. isolates carrying antimicrobial resistance genes recovered from patients in Kenya with acute or severe diarrheal disease. The phylogenetic tree was constructed by using a maximum-likelihood single-nucleotide polymorphism core genome alignment with a reference strain. Isolates are identified by reference genome identification numbers. Tree scale bar measures substitutions per site. AMR, antimicrobial resistance; AST, antimicrobial susceptibility testing; AZM, azithromycin; CIP, ciprofloxacin; LEV, levofloxacin; ST, sequence type.
Page created: October 16, 2024
Page updated: November 11, 2024
Page reviewed: November 11, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.