Volume 30, Number 2—February 2024
Research
Piscichuvirus-Associated Severe Meningoencephalomyelitis in Aquatic Turtles, United States, 2009–2021
Figure 4
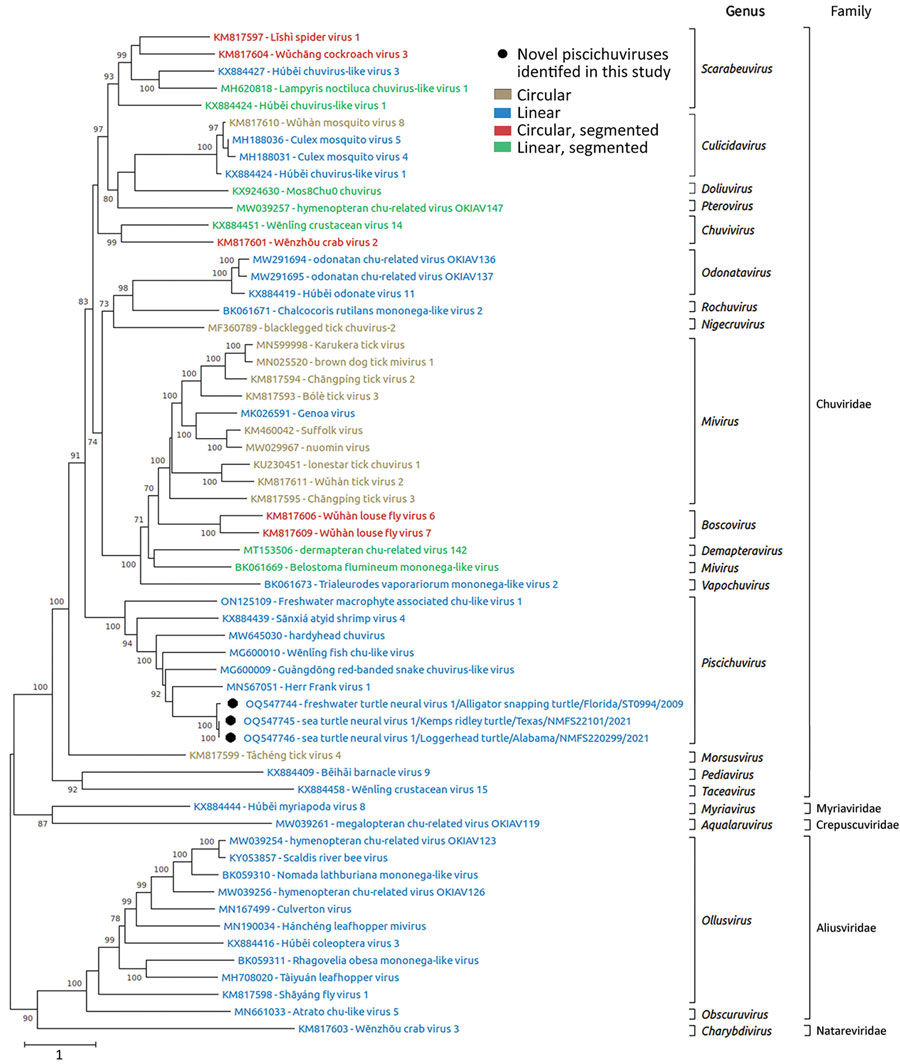
Figure 4. Phylogenetic analysis of jingchuviral large protein (L) amino acid sequences from piscichuvirus-infected aquatic turtles with meningoencephalomyelitis, United States, 2009–2021 (black dots), and reference sequences. Complete L amino acid sequences were aligned by using ClustalW (https://www.clustal.org) and refined by using MUSCLE (https://www.ebi.ac.uk/Tools/msa/muscle) with default settings. The phylogenetic analysis was performed on MEGA X (23) by using the maximum-likelihood method and Le Gascuel matrix plus observed amino acid frequencies plus 5 discrete gamma categories distribution with parameter of 1.0728 plus invariant sites with 0.65% sites. The substitution model was constructed with 500 bootstrap replicates. The tree is drawn to scale; bootstrap values are measured in the number of substitutions per site. This analysis included 59 aa sequences. Sequences are color coded based on their genomic structure.
References
- Senko JF, Burgher KM, Del Mar Mancha-Cisneros M, Godley BJ, Kinan-Kelly I, Fox T, et al. Global patterns of illegal marine turtle exploitation. Glob Change Biol. 2022;28:6509–23. DOIPubMedGoogle Scholar
- Rhodin AGJ, Iverson JB, Bour R, Fritz U, Georges A, Shaffer HB, et al.; Turtle Taxonomy Working Group. Turtles of the world: annotated checklist and atlas of taxonomy, synonymy, distribution, and conservation status. In: Rhodin AGJ, Iverson JB, van Dijk PP, Stanford CB, Goode EV, Buhlmann, KA, et al., editors. Conservation Biology of Freshwater Turtles and Tortoises: a Compilation Project of the IUCN/SSC Tortoise and Freshwater Turtle Specialist Group. 9th ed. Chelonian Research Monographs. Rochester (NY): Mercury Print Productions. 2021;8:1–472.
- Greenblatt RJ, Work TM, Dutton P, Sutton CA, Spraker TR, Casey RN, et al. Geographic variation in marine turtle fibropapillomatosis. J Zoo Wildl Med. 2005;36:527–30. DOIPubMedGoogle Scholar
- Waltzek TB, Stacy BA, Ossiboff RJ, Stacy NI, Fraser WA, Yan A, et al. A novel group of negative-sense RNA viruses associated with epizootics in managed and free-ranging freshwater turtles in Florida, USA. PLoS Pathog. 2022;18:
e1010258 . DOIPubMedGoogle Scholar - Harding EF, Russo AG, Yan GJH, Mercer LK, White PA. Revealing the uncharacterised diversity of amphibian and reptile viruses. ISME Commun. 2022;2:95. DOIPubMedGoogle Scholar
- Li CX, Shi M, Tian JH, Lin XD, Kang YJ, Chen LJ, et al. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. eLife. 2015;4:1–26. DOIPubMedGoogle Scholar
- Han X, Wang H, Wu N, Liu W, Cao M, Wang X. Leafhopper Psammotettix alienus hosts chuviruses with different genomic structures. Virus Res. 2020;285:
197992 . DOIPubMedGoogle Scholar - Shi M, Lin XD, Tian JH, Chen LJ, Chen X, Li CX, et al. Redefining the invertebrate RNA virosphere. Nature. 2016;540:539–43. DOIPubMedGoogle Scholar
- Shi M, Lin XD, Chen X, Tian JH, Chen LJ, Li K, et al. The evolutionary history of vertebrate RNA viruses. Nature. 2018;556:197–202. DOIPubMedGoogle Scholar
- Hahn MA, Rosario K, Lucas P, Dheilly NM. Characterization of viruses in a tapeworm: phylogenetic position, vertical transmission, and transmission to the parasitized host. ISME J. 2020;14:1755–67. DOIPubMedGoogle Scholar
- Dezordi FZ, Vasconcelos CRDS, Rezende AM, Wallau GL. In and outs of Chuviridae endogenous viral elements: origin of a potentially new retrovirus and signature of ancient and ongoing arms race in mosquito genomes. Front Genet. 2020;11:
542437 . DOIPubMedGoogle Scholar - Di Paola N, Dheilly NM, Junglen S, Paraskevopoulou S, Postler TS, Shi M, et al. Jingchuvirales: a new taxonomical framework for a rapidly expanding order of unusual monjiviricete viruses broadly distributed among arthropod subphyla. Appl Environ Microbiol. 2022;88:
e0195421 . DOIPubMedGoogle Scholar - Argenta FF, Hepojoki J, Smura T, Szirovicza L, Hammerschmitt ME, Driemeier D, et al. Identification of reptarenaviruses, hartmaniviruses and a novel chuvirus in captive Brazilian native boa constrictors with boid inclusion body disease. J Virol. 2020;94:1–19. DOIPubMedGoogle Scholar
- Conceição-Neto N, Yinda KC, Van Ranst M, Matthijnssens J. NetoVIR: modular approach to customize sample preparation procedures for viral metagenomics. In: Moya A, Pérez Brocal V, editors. The Human Virome In Molecular Biology. Totowa (NJ): Humana Press Inc.; 2018. p. 85–95.
- Vibin J, Chamings A, Collier F, Klaassen M, Nelson TM, Alexandersen S. Metagenomics detection and characterisation of viruses in faecal samples from Australian wild birds. Sci Rep. 2018;8:8686. DOIPubMedGoogle Scholar
- Young KT, Stephens JQ, Poulson RL, Stallknecht DE, Dimitrov KM, Butt SL, et al. Putative novel avian paramyxovirus (AMPV) and reidentification of APMV-2 and APMV-6 to the species level based on wild bird surveillance (United States, 2016–2018). Appl Environ Microbiol. 2022;88:
e0046622 . DOIPubMedGoogle Scholar - Young KT, Lahmers KK, Sellers HS, Stallknecht DE, Poulson RL, Saliki JT, et al. Randomly primed, strand-switching, MinION-based sequencing for the detection and characterization of cultured RNA viruses. J Vet Diagn Invest. 2021;33:202–15. DOIPubMedGoogle Scholar
- Kim D, Song L, Breitwieser FP, Salzberg SL. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 2016;26:1721–9. DOIPubMedGoogle Scholar
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37:540–6. DOIPubMedGoogle Scholar
- Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. DOIPubMedGoogle Scholar
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. DOIPubMedGoogle Scholar
- Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–31. DOIPubMedGoogle Scholar
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. DOIPubMedGoogle Scholar
- VanDevanter DR, Warrener P, Bennett L, Schultz ER, Coulter S, Garber RL, et al. Detection and analysis of diverse herpesviral species by consensus primer PCR. J Clin Microbiol. 1996;34:1666–71. DOIPubMedGoogle Scholar
- Byles RA. Behavior and ecology of sea turtles from Chesapeake Bay, Behavior and ecology of sea turtles from Chesapeake Bay, Virginia [dissertation]. Williamsburg (VA): College of William and Mary; 1988.
- Jackson GJ Jr, Ross A. The occurrence of barnacles on the alligator snapping turtle, Macrochelys temminckii (Troost). J Herpetol. 1971;5:188–9. DOIGoogle Scholar
- Obijeski JF, McCauley J, Skehel JJ. Nucleotide sequences at the terminal of La Crosse virus RNAs. Nucleic Acids Res. 1980;8:2431–8. DOIPubMedGoogle Scholar
- Hsu MT, Parvin JD, Gupta S, Krystal M, Palese P. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc Natl Acad Sci U S A. 1987;84:8140–4. DOIPubMedGoogle Scholar
- Auperin DD, Romanowski V, Galinski M, Bishop DH. Sequencing studies of pichinde arenavirus S RNA indicate a novel coding strategy, an ambisense viral S RNA. J Virol. 1984;52:897–904. DOIPubMedGoogle Scholar
- Fodor E, Pritlove DC, Brownlee GG. The influenza virus panhandle is involved in the initiation of transcription. J Virol. 1994;68:4092–6. DOIPubMedGoogle Scholar
- Neumann G, Hobom G. Mutational analysis of influenza virus promoter elements in vivo. J Gen Virol. 1995;76:1709–17. DOIPubMedGoogle Scholar
- Flick R, Neumann G, Hoffmann E, Neumeier E, Hobom G. Promoter elements in the influenza vRNA terminal structure. RNA. 1996;2:1046–57.PubMedGoogle Scholar
- Keatts LO, Robards M, Olson SH, Hueffer K, Insley SJ, Joly DO, et al. Implications of zoonoses from hunting and use of wildlife in North American arctic and boreal biomes: pandemic potential, monitoring, and mitigation. Front Public Health. 2021;9:
627654 . DOIPubMedGoogle Scholar - Fredricks DN, Relman DA. Sequence-based identification of microbial pathogens: a reconsideration of Koch’s postulates. Clin Microbiol Rev. 1996;9:18–33. DOIPubMedGoogle Scholar
- Heppell SS. Application of life-history theory and population model analysis to turtle conservation. Copeia. 1998;1998:367–75. DOIGoogle Scholar
- Thomas TM, Granatosky MC, Bourque JR, Krysko KL, Moler PE, Gamble T, et al. Taxonomic assessment of Alligator Snapping Turtles (Chelydridae: Macrochelys), with the description of two new species from the southeastern United States. Zootaxa. 2014;3786:141–65. DOIPubMedGoogle Scholar
- Folt B, Guyer C. Evaluating recent taxonomic changes for alligator snapping turtles (Testudines: Chelydridae). Zootaxa. 2015;3947:447–50. DOIPubMedGoogle Scholar