Volume 30, Number 4—April 2024
Research
Emergence of Poultry-Associated Human Salmonella enterica Serovar Abortusovis Infections, New South Wales, Australia
Figure 3
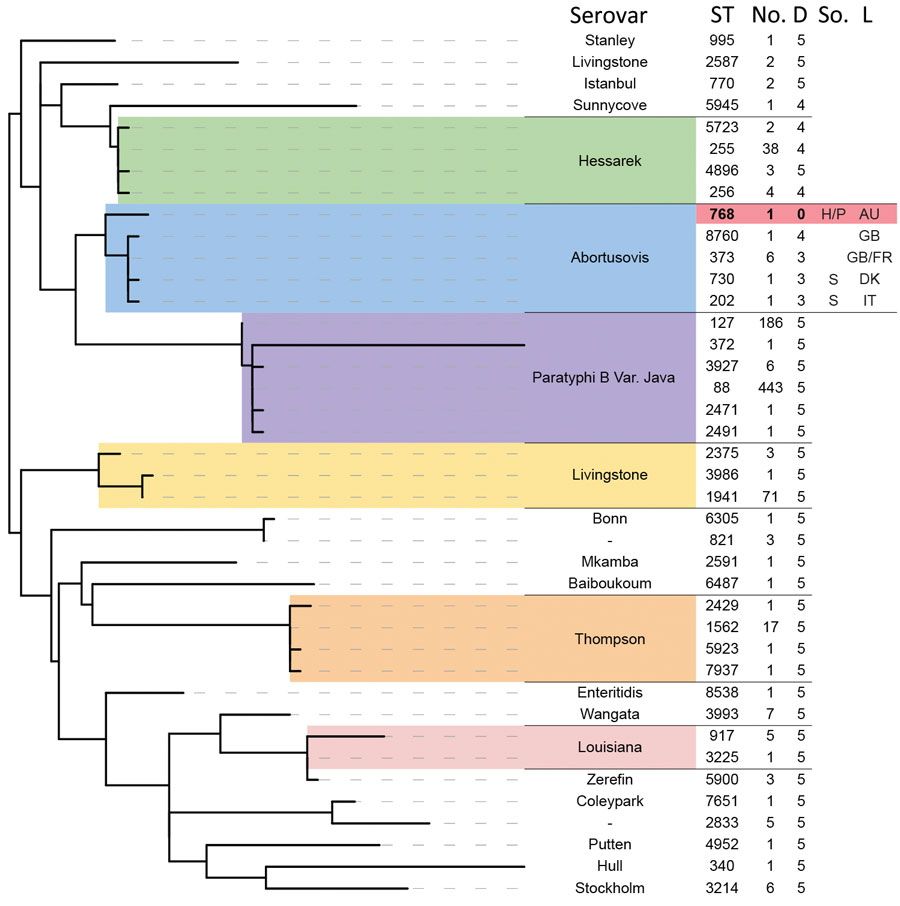
Figure 3. Phylogenetic relationship of other serovars to isolates in a study of the emergence of poultry-associated human Salmonella enterica serovar Abortusovis infections, New South Wales, Australia using MLST sequence data. Sequence types in the Enterobase database with five or fewer allele differences from the Australian ST768 (shaded in red) were identified and used to generate a phylogeny of related Salmonella isolates using maximum likelihood method. All branches with less than 50% bootstrap support were collapsed. The ST at each terminal branch is shown, as is the number of isolates in the Enterobase database assigned that ST and the number of allele differences from ST768. Available source and location data for Salmonella Abortusovis STs are displayed. Two letter abbreviations are used for country of origin. AU, Australia; D, allele difference from ST 768; DK, Denmark; FR, France; GB, Great Britain; H, human; IT, Italy; L, location; P, poultry; S, sheep; So., source; ST, sequence type.