Volume 30, Number 6—June 2024
Dispatch
Zoonotic Ancylostoma ceylanicum Infection in Coyotes from Guanacaste Conservation Area, Costa Rica, 2021
Figure 1
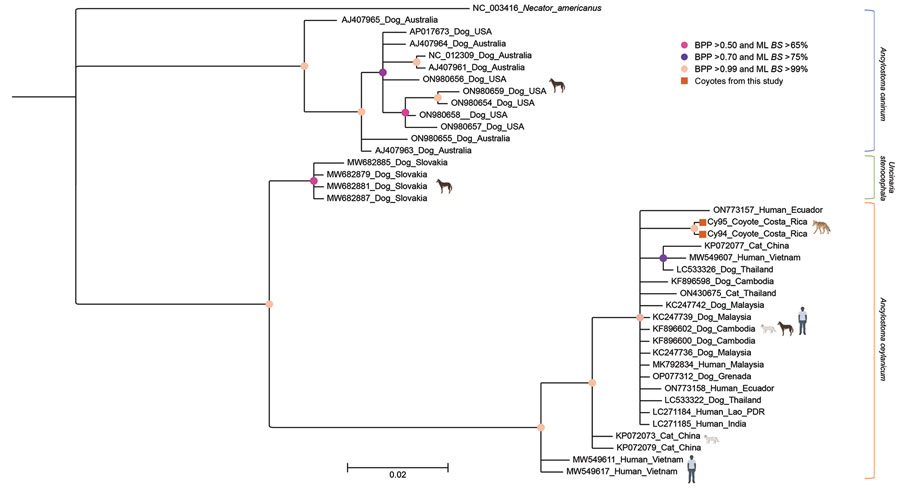
Figure 1. Phylogeny of Ancylostoma ceylanicum hookworms from coyotes in from Guanacaste Conservation Area, Costa Rica, 2021, and reference sequences. Gene tree shows a partial region of the mitochondrial cox1 gene for the hookworms A. caninum, A. ceylanicum, and Uncinaria stenocephala. Phylogenetic analyses were conducted using Bayesian inference in MrBayes v3.2.7a, employing the Hasegawa-Kishino-Yano substitution model with 4 gamma-distributed categories. Four independent Markov chains ran for 1,000,000 Markov chain Monte Carlo generations, with tree sampling occurring every 200 generations. The initial 25% of generated trees was considered burn-in, while the remaining trees were used to derive consensus trees. The analysis proceeded until the potential scale reduction factor approached 1, and the average SD of split frequencies was <0.01. For maximum-likelihood analysis, we used the rapid bootstrapping option with 10,000 iterations based on the Akaike information criterion. To root the tree, we included the human hookworm Necator americanus (GenBank NC_003416). Circles on nodes indicate BPP and ML BS percentages. Taxon names are annotated with GenBank accession numbers, host, and country of origin. Scale bar indicates the mean number of nucleotide substitutions per site. BPP, Bayesian posterior probability; ML BS, maximum-likelihood bootstrap support.