Volume 30, Number 6—June 2024
Research Letter
Novel Avian Influenza A(H5N6) Virus in Wild Birds, South Korea, 2023
Figure
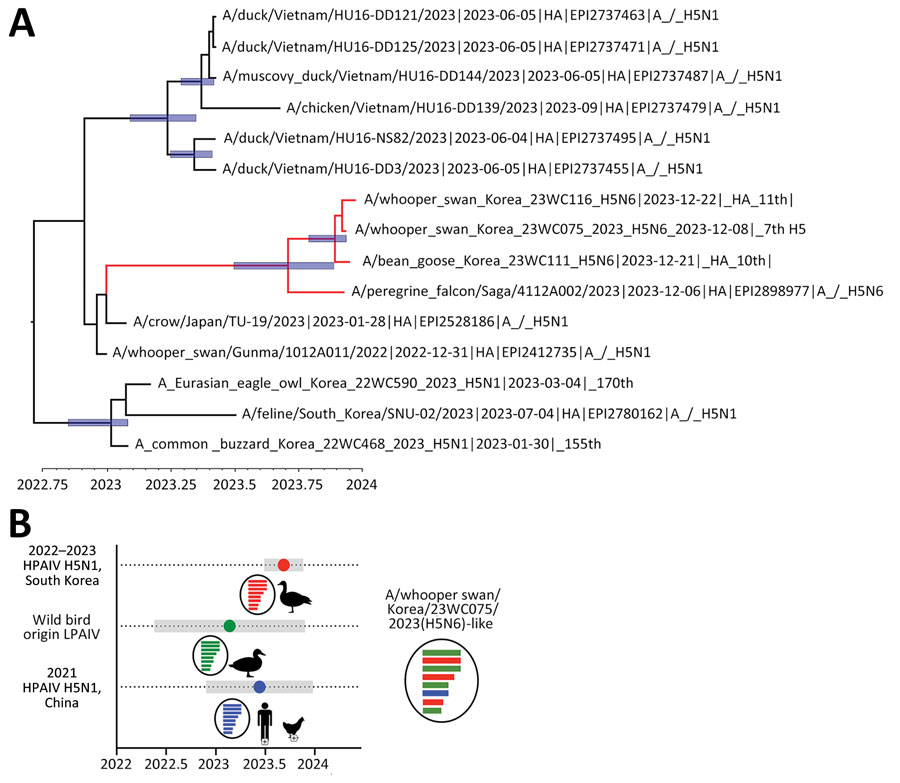
Figure. Exploration of most common ancestors for novel reassortant highly pathogenic avian influenza A(H5N6) clade 2.3.4.4 b isolates recovered from wild birds, South Korea. A) Maximum clade credibility tree of viruses found in the carcasses of whooper swans and bean geese in South Korea, December 2023. Tree was constructed using the hemagglutinin gene of the H5N6 viruses. Red indicates H5N6 isolates from South Korea and Japan. The timescale is shown on the horizontal axis in decimal years. Node bars represent 95% highest posterior density of the heights. Accession numbers beginning with EPI indicate isolates from the GISAID database (https://www.gisaid.org). B) Temporal schematic of the reassortant genome constellation of the novel reassortant H5N6 viruses from South Korea. Gene segments originating from H5N1 HPAIVs (red), LPAIVs (green), and H5N6 (blue) HPAIVs are indicated. Shade bars represent 95% highest posterior density range of time to most recent common ancestor. Circles represent the mean time to most recent common ancestor. HPAIV, highly pathogenic avian influenza virus; LPAIV, low-pathogenicity avian influenza viruses.
1These authors contributed equally to this article.