Group B Streptococcus Sequence Type 103 as Human and Bovine Pathogen, Brazil
Laura M.A. Oliveira

, Leandro C. Simões, Chiara Crestani, Natália S. Costa, José Carlos F. Pantoja, Renata F. Rabello, Sérgio E.L. Fracalanzza, Lucia M. Teixeira, Uzma B. Khan, Dorota Jamrozy, Stephen Bentley, Tatiana C.A. Pinto, and Ruth N. Zadoks
Author affiliations: Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (L.M.A. Oliveira, L.C. Simões, N.S. Costa, S.E.L. Fracalanzza, L.M. Teixeira, T.C.A. Pinto); Pasteur Institut, Paris, France (C. Crestani); Universidade Estadual Paulista Júlio de Mesquita Filho, São Paulo, Brazil (J.C.F. Pantoja); Fluminense Federal University, Rio de Janeiro (R.F. Rabello); Wellcome Sanger Institute, Hinxton, UK (U.B. Khan, D. Jamrozy, S. Bentley); University of Sydney, Camden, New South Wales, Australia (R.N. Zadoks)
Main Article
Figure 2
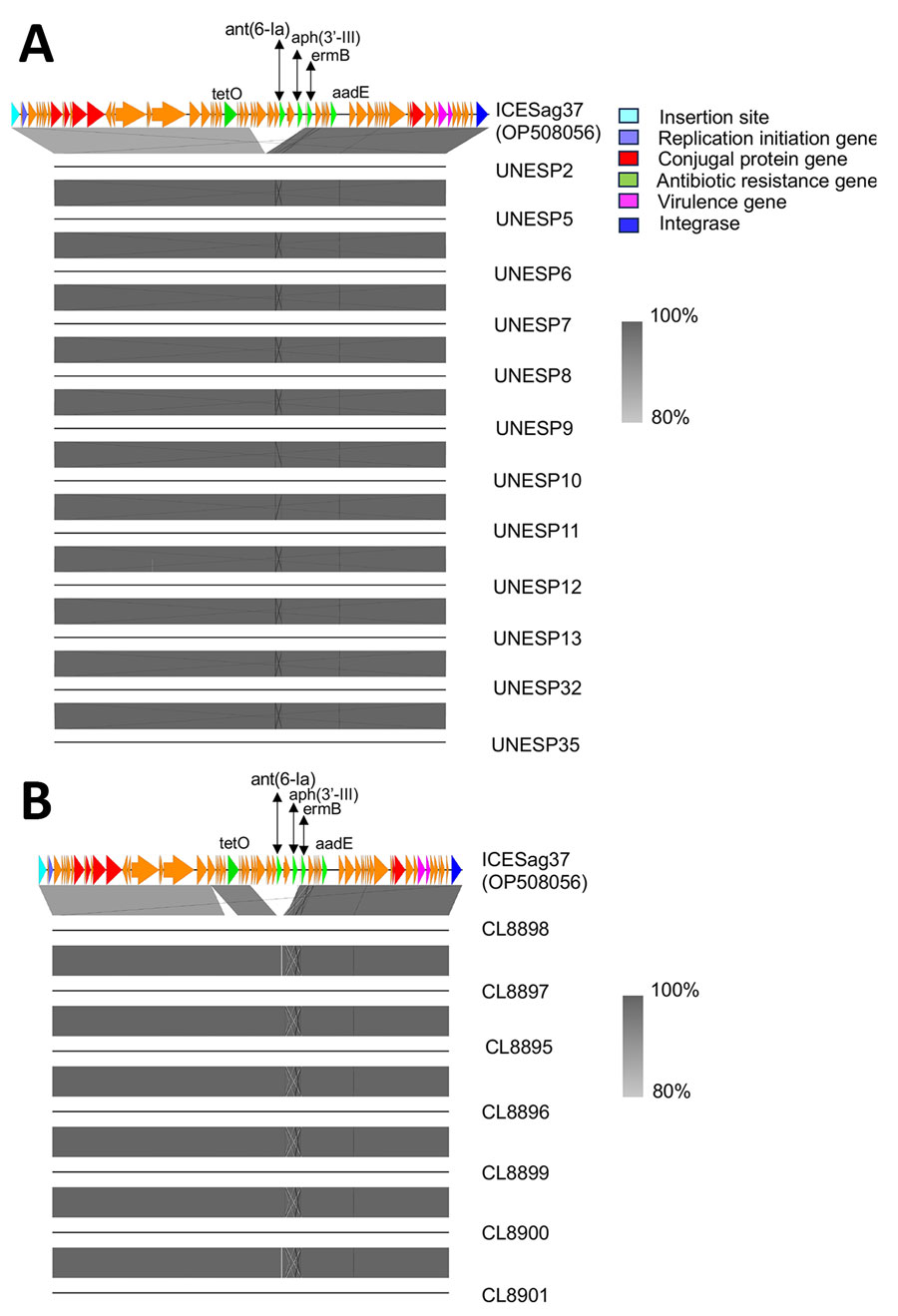
Figure 2. Alignment of nucleotide sequences of bovine group B Streptococcus isolates belonging to ST103 collected in Brazil with ICESag37, an integrative conjugative element associated with antimicrobial resistance and virulence genes. A) Sequence alignment for cluster D isolates (n = 12). B) Sequence alignment for cluster E isolates (n = 7). The analysis used Easyfig version 2.2 to perform BLASTn (https://blast.ncbi.nlm.nih.gov) comparisons between isolates in clusters D and E against the reference ICEsag37 (GenBank accession no. OP508056). Arrows indicate mobility genes and conjugal transfer proteins (red), repA initiator genes (purple), critical site-specific recombinase (blue), antibiotic resistance genes (green), and other ICE genes (orange). Aminoglycoside genes normally found in ICESag37 (ant(6–1a) and aph(3-III)) are missing from the bovine isolates.
Main Article
Page created: July 10, 2024
Page updated: July 21, 2024
Page reviewed: July 21, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
