Volume 30, Number 8—August 2024
Dispatch
Macrolide-Resistant Mycoplasma pneumoniae Infections among Children before and during COVID-19 Pandemic, Taiwan, 2017–2023
Cite This Article
Citation for Media
Abstract
Before the COVID-19 pandemic, Mycoplasma pneumoniae infections emerged during spring to summer yearly in Taiwan, but infections were few during the pandemic. M. pneumoniae macrolide resistance soared to 85.7% in 2020 but declined to 0% during 2022–2023. Continued molecular surveillance is necessary to monitor trends in macrolide-resistant M. pneumoniae.
Mycoplasma pneumoniae is a major cause of respiratory tract infections, particularly in children and young adults. M. pneumoniae accounts for 10%–30% of community-acquired pneumonia (1). Macrolides are the primary treatment, but since 2000, macrolide-resistant M. pneumoniae (MRMP) strains have increased substantially, especially in Asia (2,3). However, a global study indicated a decline in M. pneumoniae detections during 2017–2021, which researchers attributed to the impact of nonpharmaceutical COVID-19 measures on M. pneumoniae transmission (4).
In Taiwan, MRMP prevalence rose from 12.3%–24% before 2017 to 54%–88% during 2017–2020 (5–7). Multilocus sequence typing (MLST) is a valuable tool for epidemiologic surveillance, offering high discriminatory ability to identify the shift of circulating strain types (8–10). We used MLST to analyze the genetic diversity and macrolide resistance prevalence of M. pneumoniae in hospitalized children in Taiwan during 2017–2023. This study was approved by the institutional review board of Show Chwan Memorial Hospital (approval no 1051007 and 1091104).
Our study spanned 2 phases: phase 1 was March 2017–June 2019, and phase 2 was March 2020–December 2023. After obtaining the necessary consent, we enrolled children <18 years of age at 4 central hospitals in Taiwan: Show Chwan Memorial Hospital (739 beds), Chang Bing Show Chwan Memorial Hospital (946 beds), Jen-Ai Hospital (644 beds), and Chung Shan Medical University Hospital (1,023 beds). We enrolled children with acute respiratory tract infections who tested M. pneumoniae IgM–positive via Biocardä (AniBiotech, https://anibiotech.fi). We collected nasopharyngeal or oropharyngeal swab specimens, stored them, and cultured for M. pneumoniae. We performed MLST typing on positive cultures confirmed by PCR as true M. pneumoniae infections.
We cultured specimens in SP-4 medium and incubated at 37°C in 5% CO2 for 14 days (11). We used the QIAamp DNA Blood Mini Kit (QIAGEN, https://www.qiagen.com) to extract DNA and confirmed M. pneumoniae by real-time PCR targeting the repMp1 gene (12). We further analyzed M. pneumoniae isolates, including single-base mutations in the 23S rRNA gene and MLST on the basis of 8 housekeeping genes (8). We used goeBURST software (https://phyloviz.readthedocs.io/en/latest/data_analysis.html) to explore relationships among sequence types (STs).
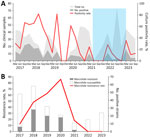
Among 770 nasopharyngeal or oropharyngeal samples collected during the study period, 209 (27.1%) were confirmed M. pneumoniae cases. By comparing the annual and seasonal distribution of M. pneumoniae respiratory tract infections, we noted significant yearly differences (p<0.001) and an outbreak that occurred during 2017–2018 (Table 1). Seasonal prevalence also varied significantly (p<0.001); average positivity rates were 30% in spring, 41.6% in summer, 18.1% in autumn, and 22.1% in winter. M. pneumoniae was detected year-round, but primarily during March–August (Figure 1, panel A). During the COVID-19 pandemic, detection rates notably declined.
Among the 209 M. pneumoniae–positive isolates, 74 (35.4%) were macrolide-resistant. Annual resistance rates ranged from 12.5% in 2017 to 85.7% in 2020, then dropped to 18.2% in 2021 and 0% in 2022 and 2023 (Figure 1, panel B). Among macrolide-resistant isolates, 72 (97.3%) had the A2063G mutation, and 2 (2.7%) had the A2063T mutation.
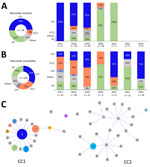
We submitted all 209 confirmed M. pneumoniae strains for molecular analysis, and 155 were identified by MLST, revealing 12 different STs (Appendix Table). ST3 was the most prevalent at 38.2%, followed by ST17 at 19.1%. However, 54 (25.8%) strains could not be successfully genotyped due to the failure of amplification and sequencing of the pgm locus. Of the 74 macrolide-resistant isolates, the leading STs were ST3 (49%) and ST17 (8%), but ST3 was the most common in MRMP during 2017–2019 (Figure 2, panel A). Among 135 macrolide-susceptible isolates, ST33 (33%), ST17 (25%), and ST9 (3%) were predominant (Figure 2, panel B). ST17 was more common in macrolide-susceptible than in macrolide-resistant isolates (p = 0.003), and ST3 was more prevalent in macrolide-resistant isolates (p = 0.002). However, we found no correlation between the dynamic proportion of macrolide resistance and ST3, although a notable percentage of macrolide-resistant strains within ST3 were detected during 2017–2019 (Table 2).
We identified 2 clonal complexes (CCs) in the goeBURST analysis (Figure 2, panel C), which included data from 140 strains. Of those 2 CC clusters, most (96%) STs belonged to CC1, which included the most frequently detected STs, ST3 and ST17. CC2 comprised 5 (4%) strains and included 2 STs, ST33 and ST26.
During 2017–2023, we observed changes in M. pneumoniae infection rates, alterations in STs, and shifts in antimicrobial resistance. Nonpharmaceutical interventions during the pandemic mitigated the transmission of respiratory pathogens besides SARS-CoV-2 (13). In our study, we noted a substantial decrease in M. pneumoniae detection during 2021, and no positive cases were recorded during June 2021–February 2022, coinciding with the height of the pandemic period in Taiwan, a finding that aligns with those reported in a global survey (4).
MRMP infections have increased greatly worldwide, particularly in the Western Pacific region (14). In Taiwan, prior studies noted a substantial rise in macrolide resistance rates from 12%–24% during 2011–2016 to 54%–88% during 2017–2020 (5–7). However, in our study, MRMP prevalence decreased rapidly to 0%–18.2% during 2021–2023. During the 2011–2012 outbreak in Japan, the MRMP detection rate soared to 90% (9). After that outbreak, the number of MRMP strains decreased, reaching 14.3% in 2018, whereas China and South Korea continued to report high resistance rates from 2014 to 2018 (10,15).
ST3 within CC1 is a globally successful clone and was prevalent in Japan, China, South Korea, Cuba, Germany, and Taiwan at rates from 30% to 70% during 2002–2022 (Appendix Figure). A previous study in Taiwan showed high macrolide resistance rates for ST3 (93.5%) and ST17 (82.1%) (7). In contrast, ST17 was more common among the macrolide-susceptible cases in our study and had a resistance rate of only 15%. Recent data indicate shifts in M. pneumoniae sequence types: in Japan, ST3 and ST14 were largely replaced by ST7 and ST33 in 2018–2019, reducing macrolide resistance by 11.3% (9). However, South Korea did not have a notable decrease in its macrolide resistance rate (78.5%) during 2019–2020, and ST3 remained dominant (10). Our findings show no substantial shifts in ST distribution in Taiwan but a notable change in the ratio of macrolide-resistant to macrolide-susceptible within ST3, possibly reducing the overall resistance rate (Table 2).
The first limitation of our study is that an 8-month gap in data collection occurred during July 2019–February 2020, which might have caused a slight underestimation of cases. Second, we only enrolled patients who tested positive for M. pneumoniae IgM, which might underestimate the actual number of M. pneumoniae infections. We did not investigate the clinical manifestations in patients who tested IgM-negative. Third, although the trend toward resistant strains was evident, the absolute number of resistant strains was small. Fourth, some strains failed at the pgm locus, and on the basis of other known loci, those strains are most likely to be either ST3 or ST17. That issue might have been to the result of low bacterial loads and fragile DNA that hindered the successful amplification of the 1,072-bp pgm locus via nested PCR. Recent advances have made whole-genome sequencing a promising tool for future M. pneumoniae research, especially in identifying genotypes linked to macrolide resistance or virulence.
In conclusion, M. pneumoniae respiratory tract infections in Taiwan exhibit seasonality, and prevalence decreased after the COVID-19 pandemic. Simultaneously, MRMP incidence experienced a sharp decline after 2021. Although ST3 remains the most prevalent M. pneumoniae strain and is associated with macrolide resistance, global data still lack evidence of correlation between STs and macrolide resistance. Thus, continued molecular surveillance is necessary to monitor those trends.
Dr. Wu is a pediatrician in Show Chwan Memorial Hospital, Changhua, Taiwan. His primary research interest is infectious diseases, particularly Mycoplasma pneumoniae.
Acknowledgment
We thank the patients who graciously consented to participate in this study, as well as all the clinicians who contributed by taking samples.
References
- Principi N, Esposito S, Blasi F, Allegra L; Mowgli study group. Role of Mycoplasma pneumoniae and Chlamydia pneumoniae in children with community-acquired lower respiratory tract infections. Clin Infect Dis. 2001;32:1281–9. DOIPubMedGoogle Scholar
- Morozumi M, Takahashi T, Ubukata K. Macrolide-resistant Mycoplasma pneumoniae: characteristics of isolates and clinical aspects of community-acquired pneumonia. J Infect Chemother. 2010;16:78–86. DOIPubMedGoogle Scholar
- Pereyre S, Goret J, Bébéar C. Mycoplasma pneumoniae: current knowledge on macrolide resistance and treatment. Front Microbiol. 2016;7:974. DOIPubMedGoogle Scholar
- Meyer Sauteur PM, Beeton ML, Uldum SA, Bossuyt N, Vermeulen M, Loens K, et al.; ESGMAC–MyCOVID Study Team. Mycoplasma pneumoniae detections before and during the COVID-19 pandemic: results of a global survey, 2017 to 2021. Euro Surveill. 2022;27:
2100746 . DOIPubMedGoogle Scholar - Wu HM, Wong KS, Huang YC, Lai SH, Tsao KC, Lin YJ, et al. Macrolide-resistant Mycoplasma pneumoniae in children in Taiwan. J Infect Chemother. 2013;19:782–6. DOIPubMedGoogle Scholar
- Kuo CY, Tsai WC, Lee HF, Ho TS, Huang LM, Shen CF, et al.; Taiwan Pediatric Infectious Disease Alliance (TPIDA). The epidemiology, clinical characteristics, and macrolide susceptibility of Mycoplasma pneumoniae pneumonia in children in Southern Taiwan, 2019-2020. J Microbiol Immunol Infect. 2022;55:611–9. DOIPubMedGoogle Scholar
- Hung H-M, Chuang C-H, Chen Y-Y, Liao W-C, Li S-W, Chang IY-F, et al. Clonal spread of macrolide-resistant Mycoplasma pneumoniae sequence type-3 and type-17 with recombination on non-P1 adhesin among children in Taiwan. Clin Microbiol Infect. 2021;27:1169.e1–6. DOIPubMedGoogle Scholar
- Brown RJ, Holden MT, Spiller OB, Chalker VJ. Development of a multilocus sequence typing scheme for molecular typing of Mycoplasma pneumoniae. J Clin Microbiol. 2015;53:3195–203. DOIPubMedGoogle Scholar
- Morozumi M, Tajima T, Sakuma M, Shouji M, Meguro H, Saito K, et al. Sequence type changes associated with decreasing macrolide-resistant Mycoplasma pneumoniae, Japan. Emerg Infect Dis. 2020;26:2210–3. DOIPubMedGoogle Scholar
- Lee JK, Choi YY, Sohn YJ, Kim KM, Kim YK, Han MS, et al. Persistent high macrolide resistance rate and increase of macrolide-resistant ST14 strains among Mycoplasma pneumoniae in South Korea, 2019-2020. J Microbiol Immunol Infect. 2022;55:910–6. DOIPubMedGoogle Scholar
- Tully JG. New laboratory techniques for isolation of Mycoplasma pneumoniae. Yale J Biol Med. 1983;56:511–5.PubMedGoogle Scholar
- Dumke R, Schurwanz N, Lenz M, Schuppler M, Lück C, Jacobs E. Sensitive detection of Mycoplasma pneumoniae in human respiratory tract samples by optimized real-time PCR approach. J Clin Microbiol. 2007;45:2726–30. DOIPubMedGoogle Scholar
- Mendez-Brito A, El Bcheraoui C, Pozo-Martin F. Systematic review of empirical studies comparing the effectiveness of non-pharmaceutical interventions against COVID-19. J Infect. 2021;83:281–93. DOIPubMedGoogle Scholar
- Kim K, Jung S, Kim M, Park S, Yang HJ, Lee E. Global trends in the proportion of macrolide-resistant Mycoplasma pneumoniae infections: a systematic review and meta-analysis. JAMA Netw Open. 2022;5:
e2220949 . DOIPubMedGoogle Scholar - Zhao F, Li J, Liu J, Guan X, Gong J, Liu L, et al. Antimicrobial susceptibility and molecular characteristics of Mycoplasma pneumoniae isolates across different regions of China. Antimicrob Resist Infect Control. 2019;8:143. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: July 16, 2024
Table of Contents – Volume 30, Number 8—August 2024
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Chun-Yi Lee, Department of Pediatrics, Chang Bing Show Chwan Memorial Hospital, No. 6 Lu-Kung Rd, Chang Bing Industrial Center, Lu-Kang, Changhua 505, Taiwan
Top