Volume 30, Number 8—August 2024
Dispatch
Real-Time Enterovirus D68 Outbreak Detection through Hospital Surveillance of Severe Acute Respiratory Infection, Senegal, 2023
Figure 3
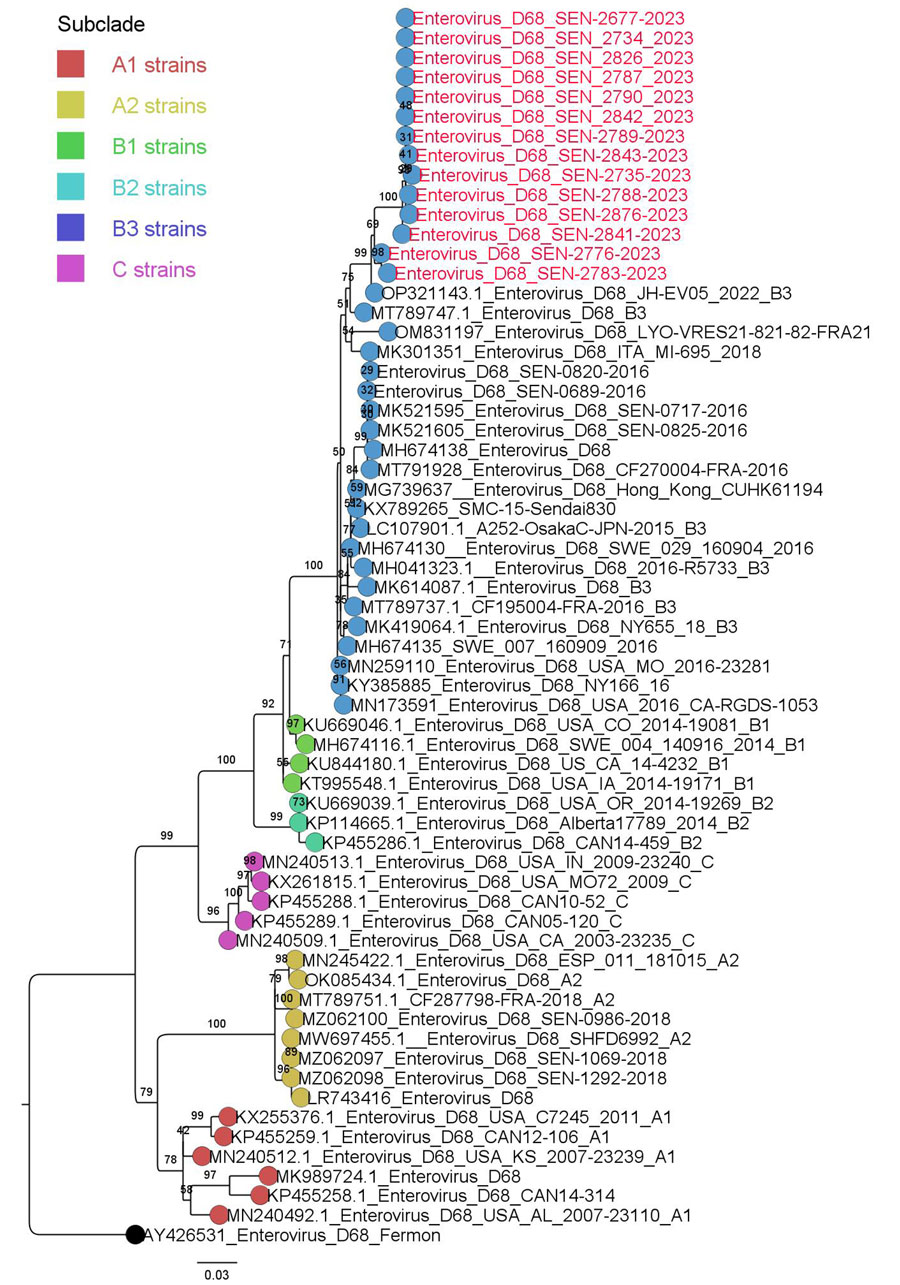
Figure 3. Maximum-likelihood phylogenetic tree based on the nucleotide sequences of major capsid protein gene region of enterovirus D68 from Senegal (red text) and reference sequences. Tree was constructed by using IQ-TREE2 2.0.6 (http://www.iqtree.org) and visualized by using Figtree 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree). Statistical significance was tested by using 1,000 bootstrapping replicates. Software was used to define the correct model used. Tree is rooted by the Fermon strain. Scale bar indicates substitutions per site.
Page created: July 03, 2024
Page updated: July 21, 2024
Page reviewed: July 21, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.