Volume 31, Number 1—January 2025
Research Letter
Salmonella enterica Serovar Abony Outbreak Caused by Clone of Reference Strain WDCM 00029, Chile, 2024
Figure 1
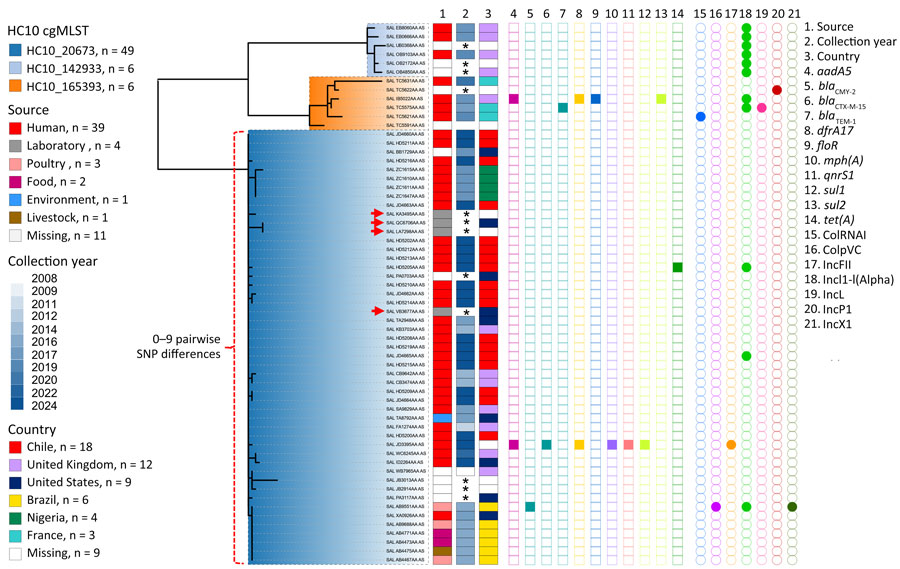
Figure 1. Phylogenetic analysis of Salmonella enterica serovar Abony from an outbreak caused by a WDCM 00029 Clone, Chile, 2024. A core SNP maximum-likelihood phylogenetic tree was constructed by using RAxML version 8 (https://github.com/stamatak/standard-RAxML) and genomes of Salmonella Abony isolates from the HC50_20673 cluster with the ATCC 6017 genome as the reference (Enterobase Barcode SAL_BA5138AA; Sequence Read Archive accession no. SRR1786283). The tree was constructed by using the Enterobase pipelines refMasker, refMapper, refMapperMatrix, and matrix_phylogeny, which together masked repeated regions, tandem repeats, and CRISPR regions in the reference genome, aligned genomes to reference, called nonrepetitive core SNPs, and built the maximum likelihood tree. Metadata regarding HC10 clusters (<10 allele differences) include isolation source, collection year, country of origin, antibiotic drug resistance genes (AMRFinderPlus version 3.12.8; database version 2024-05-02.2; https://www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance/AMRFinder), and plasmid replicons (ABRicate version 1.0.1, https://github.com/tseemann/abricate; PlasmidFinder database, updated June 4, 2024). Red arrows indicate the WDCM 00029 genomes found in the public databases (Enterobase, https://enterobase.warwick.ac.uk/species/index/senterica; National Center for Biotechnology Information Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra) that were made public in 2013 (accession no. SRR955283), 2016 (accession no. SRR1815498), 2019 (accession no. SRR8599079), and 2021 (accession no. SRR15145673). Asterisks (*) in the collection year column indicate that information was missing. The figure was made by using iTOL version 6.9 (https://itol.embl.de). ATCC, American Type Culture Collection; cgMLST, core genome multilocus sequence typing; CRISPR, clustered regularly interspaced short palindromic repeats; SNP, single-nucleotide polymorphism.