Volume 31, Number 1—January 2025
Research Letter
Salmonella enterica Serovar Abony Outbreak Caused by Clone of Reference Strain WDCM 00029, Chile, 2024
Figure 2
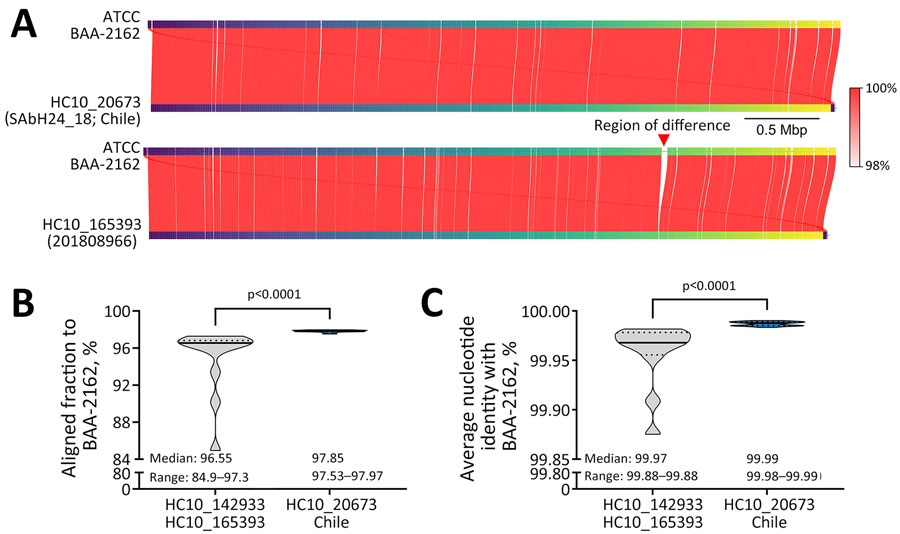
Figure 2. Whole-genome comparisons of Salmonella enterica serovar Abony from an outbreak caused by a WDCM 00029 clone, Chile, 2024. A) Example whole-genome comparison between Salmonella Abony WDCM 00029 (genome provided by ATCC; strain BAA-2162) and an isolate from the HC10_20673 cluster (strain SAbH24_18) from Chile, or the most closely related isolate (strain 201808966) outside the HC10_20673 cluster (mean difference to HC10_20673 isolates: 73 SNPs). Red lines connect regions of genome identity between each pair of compared genomes, with color indicating the percent identity. The red vertical arrow points to a region of difference between the compared genomes. B, C) Truncated violin plots of AF (B) and ANI (C) to the WDCM 00029 genome of the Salmonella Abony isolates (n = 18 genomes) from Chile and other isolates from the same HC50 cluster (HC10_142933 and HC10_165393; n = 12 genomes). In the violin plots, black horizontal lines represent medians and dotted lines represent 25% and 75% quartiles. Differences between the median values were assessed by using Mann-Whitney tests. AF and ANI calculations were made with FastANI version 1.34 (https://github.com/ParBliSS/FastANI). AF, alignment fraction; ANI, average nucleotide identity; ATCC, American Type Culture Collection; SNP, single-nucleotide polymorphism.