Volume 31, Number 1—January 2025
Dispatch
Detection and Genomic Characterization of Novel Mammarenavirus in European Hedgehogs, Italy
Figure 2
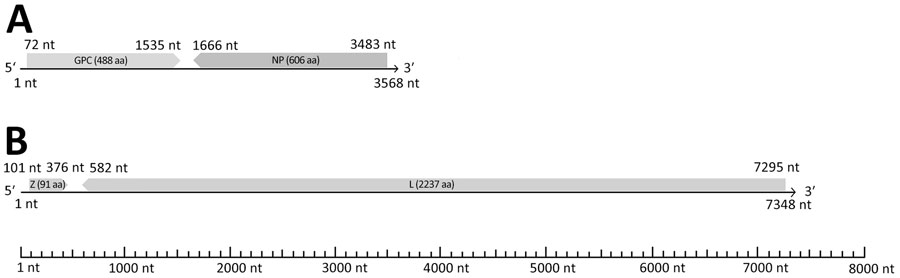
Figure 2. Schematic representation of the bisegmented genome organization of the Erinaceus Europaeus arenaviruses (EEAVs) detected in study of detection and genomic characterization of novel mammarenavirus in European hedgehogs, Italy, from 5′ to 3′ ends. A) The EEAV S genome segment (3,568 nt) coding for the putative GPC (1,464 nt) and NP (1,818 nt) proteins. B) The EEAV L genome segment (7,348 nt) encoding the putative Z (276 nt) and L proteins (6,714 nt). The proteins are shown in different shades of gray. Arrows indicate the direction of open reading frames. GPC, glycoprotein precursor; L, large; NP, nucleoprotein; S, small; Z, zinc-encoded matrix.
Page created: November 15, 2024
Page updated: December 22, 2024
Page reviewed: December 22, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.