Volume 31, Number 10—October 2025
Synopsis
Retrospective Analysis of Historical Listeria monocytogenes Clinical Isolates, New York, USA, 2000–20211
Figure 1
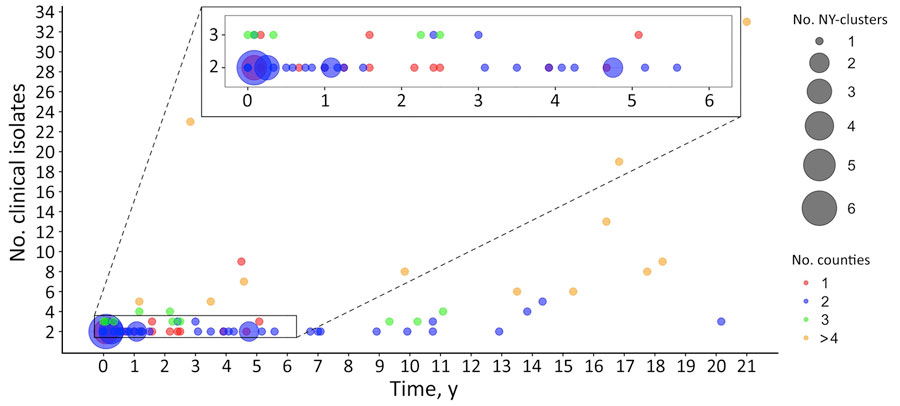
Figure 1. Distribution of timespan, number of clinical isolates, and number of counties among clusters in retrospective analysis of historical Listeria monocytogenes clinical isolates, New York, USA, 2000–2021. Clinical isolates that show a single-nucleotide polymorphism (SNP) distance of <20 SNPs to >1 of the other clinical isolates were grouped into a cluster; therefore, each cluster contains >2 clinical isolates. Each circle represents between 1 and 6 clusters, and the size of the circle is proportional to the number of clusters represented. Circles are color-coded according to the number of counties in each cluster. Timespan represents the interval between the first and the last collected clinical isolates in a cluster, measured in years with a month (1/12 of a year) as the minimum measurable unit. If the first and last collected clinical isolates in a cluster were obtained within the same month of the same year, the timespan for the cluster is zero.
1Preliminary results from this study were presented at the International Association for Food Protection Conference; July 14–17, 2024; Long Beach, California, USA.