Volume 31, Number 10—October 2025
Research Letter
Detection of Burkholderia thailandensis in Soil Samples, Suriname
Abstract
Melioidosis, caused by the highly lethal pathogen Burkholderia pseudomallei, is emerging in North and South America. We studied soil samples in Suriname to determine endemicity of Burkholderia species. B. thailandensis was isolated, but B. pseudomallei was not. A multidisciplinary approach could establish clinical and ecologic distribution of both Burholderia species in Suriname.
The epidemiology of the environmental gram-negative bacteria Burkholderia pseudomallei, the highly lethal pathogen causing melioidosis, and less lethal B. thailandensis is changing because of climatic conditions (1,2). During 2019–2022, B. pseudomallei and B. thailandensis, both part of the B. pseudomallei complex, were isolated from the environment in the continental United States (2,3). A large set of whole-genome sequence data indicates that B. pseudomallei could have been introduced from Africa to North and South America between 1650 and 1850 through the slave trade (4), and that might also be true for B. thailandensis.
Suriname, a tropical country in South America with an ethnically diverse population of <1 million inhabitants, could be a historic port of entry for Burkholderia species, especially considering B. pseudomallei is established in neighboring countries Brazil and (presumably) French Guiana (5,6). However, neither Burkholderia species has yet been identified in Suriname. We performed an environmental sampling study to determine whether B. pseudomallei or B. thailandensis could be isolated from soil samples in Suriname.
We collected soil samples in Suriname during the major wet season in May–July 2023 (https://climateknowledgeportal.worldbank.org/country/suriname/climate-data-historical) by using methods from the consensus guidelines for isolation of B. pseudomallei and our previous work in Nigeria (7,8). We selected 4 sites in the northern districts Paramaribo, Commewijne, and Nickerie and collected 100 samples at each site at a depth of 60–80 cm (Table). We determined the soil acidity and alkalinity and the salinity of air-dried mixed soil samples by measuring the pH and electrical conductivity. Next, we cultured 10 g of soil by using selective broth and agar and inspected agar plates daily for suspect isolates. We subcultured morphologically suspect isolates onto blood agar plates and screened for oxidase positivity by using oxidase strips (Sigma-Aldrich, https://www.sigmaaldrich.com) and antimicrobial resistance testing by using disks (Thermo Fisher Scientific, https://www.thermofisher.com) to assess susceptibility to amoxicillin/clavulanic acid at 30 μg (defined by the presence of an inhibition zone) and resistance to colistin at 10 μg and gentamicin at 10 μg (defined by the absence of an inhibition zone), considered an adjustment of the standard operating procedure of the consensus methods (7,9). Next, we subjected isolates matching those criteria to a real-time multiplex PCR on the LightCycler 480 II (Roche, https://www.lifescience.roche.com) and subsequent presumed B. thailandensis isolates were confirmed by using whole-genome sequencing on the NextSeq 500 platform (Illumina, https://www.illumina.com) (Appendix). We uploaded whole-genome sequences of the B. thailandensis isolates to the European Nucleotide Archive database (https://www.ebi.ac.uk/ena; project no. PRJEB79520, sample accession nos. ERS20924713–6).
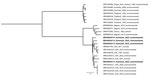
We collected 400 soil samples across 4 different sites and identified 4 B. thailandensis isolates at 1 site in the Nickerie district (Table). However, we did not detect B. pseudomallei. Sequence data revealed that the B. thailandensis isolates were phylogenetically distinct because they differed by 2,965–3,226 single-nucleotide polymorphisms from each other, suggesting multiple introductory events. We downloaded paired-end sequences of other B. thailandensis isolates with data on country and source from the European Nucleotide Archive database, including isolates identified in the continental United States and Nigeria (2,8), and constructed a phylogenetic tree (Figure). The isolates from Suriname clustered in the same clade with the isolates from the United States and Nigeria, suggesting the Suriname isolates could have descended from Africa.
We might have underestimated the true distribution of B. thailandensis and cannot rule out the presence of B. pseudomallei because of the small sample size and reliance on outdated soil-focused consensus guidelines based on soil sampling in highly endemic areas (7). Furthermore, additional water and air sampling might have yielded a higher success rate in determining endemicity for B. pseudomallei but are not yet an integral part of the adopted guidelines (7).
A strength of our study is performing an environmental sampling study in different districts in the northern parts of Suriname, where the population and agricultural practices are dense and interaction between humans and soil is highly likely. However, we did not find B. pseudomallei, which might reflect a low soil abundance. In line with this finding, a previous study of traveler-associated melioidosis cases in the Netherlands did not identify any cases that originated in Suriname (10), despite frequent family visits between Suriname and the Netherlands. Our phylogenetic results could support the theory that B. thailandensis was introduced to the Americas through slave trade, similar to the continental introduction of B. pseudomallei, suggested by others on the basis of sequence data of 469 isolates (4).
In conclusion, we detected B. thailandensis in soil samples from Suriname. Although we did not detect B. pseudomallei, a multidisciplinary approach could clarify the clinical and ecologic distribution of both Burholderia species in Suriname.
Mr. Savelkoel is a researcher at the Center for Infection and Molecular Medicine of Amsterdam UMC. His research interests include the global distribution and global health aspects of melioidosis.
Acknowledgments
We are grateful to everyone who helped with the collection, processing, and analysis of the samples. We especially thank the team of Verenigde Cultuur Maatschappijen at Rust en Werk for their kind gestures.
This work was financially supported by the Stichting Studiefonds KETEL 1 and the Cultuurfonds. J.S. was supported by an Amsterdam UMC PhD scholarship and E.B. by a Veni grant of the Dutch Research Council (reference no. 09150162210184).
References
- Meumann EM, Limmathurotsakul D, Dunachie SJ, Wiersinga WJ, Currie BJ. Burkholderia pseudomallei and melioidosis. Nat Rev Microbiol. 2024;22:155–69. DOIGoogle Scholar
- Hall CM, Stone NE, Martz M, Hutton SM, Santana-Propper E, Versluis L, et al. Burkholderia thailandensis isolated from the environment, United States. Emerg Infect Dis. 2023;29:618–21. DOIGoogle Scholar
- Petras JK, Elrod MG, Ty MC, Dawson P, O’Laughlin K, Gee JE, et al. Locally acquired melioidosis linked to environment—Mississippi, 2020–2023. N Engl J Med. 2023;389:2355–62. DOIGoogle Scholar
- Chewapreecha C, Holden MT, Vehkala M, Välimäki N, Yang Z, Harris SR, et al. Global and regional dissemination and evolution of Burkholderia pseudomallei. Nat Microbiol. 2017;2:16263. DOIGoogle Scholar
- Rolim DB, Lima RXR, Ribeiro AKC, Colares RM, Lima LDQ, Rodríguez-Morales AJ, et al. Melioidosis in South America. Trop Med Infect Dis. 2018;3:60. DOIGoogle Scholar
- Gasqué M, Guernier-Cambert V, Manuel G, Aaziz R, Terret J, Deshayes T, et al. Reassessing the distribution of Burkholderia pseudomallei outside known endemic areas using animal serological screening combined with environmental surveys: the case of Les Saintes (Guadeloupe) and French Guiana. PLoS Negl Trop Dis. 2024;18:
e0011977 . DOIGoogle Scholar - Limmathurotsakul D, Dance DA, Wuthiekanun V, Kaestli M, Mayo M, Warner J, et al. Systematic review and consensus guidelines for environmental sampling of Burkholderia pseudomallei. PLoS Negl Trop Dis. 2013;7:
e2105 . DOIGoogle Scholar - Savelkoel J, Oladele RO, Ojide CK, Peters RF, Notermans DW, Makinwa JO, et al. Presence of Burkholderia pseudomallei in soil, Nigeria, 2019. Emerg Infect Dis. 2023;29:1073–5. DOIGoogle Scholar
- Trinh TT, Hoang TS, Tran DA, Trinh VT, Göhler A, Nguyen TT, et al. A simple laboratory algorithm for diagnosis of melioidosis in resource-constrained areas: a study from north-central Vietnam. Clin Microbiol Infect. 2018;24:84.e1–4. DOIGoogle Scholar
- Birnie E, Savelkoel J, Reubsaet F, Roelofs JJTH, Soetekouw R, Kolkman S, et al. Melioidosis in travelers: an analysis of Dutch melioidosis registry data 1985–2018. Travel Med Infect Dis. 2019;32:
101461 . DOIGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: September 23, 2025
Table of Contents – Volume 31, Number 10—October 2025
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Jelmer Savelkoel, Amsterdam UMC location University of Amsterdam, Center for Infection and Molecular Medicine, Meibergdreef 9, 1105 AZ, Rm T1.0-234, Amsterdam, the Netherlands
Top