Trichosporon austroamericanum Infections among Hospitalized Patients, France, 2022–2024
Emilie Burel, Catherine Sartor, Valérie Moal, Vincent Bossi, Jacques Sevestre, Justine Solignac, Rémi Charrel, Marie Desnos-Ollivier, Stéphane Ranque, and Estelle Menu

Author affiliation: Aix-Marseille Université, Marseille, France (E. Burel); Institut Méditerranée-Infection, Marseille (E. Burel, V. Bossi); Assistance Publique Hôpitaux de Marseille, Hôpital Conception, Equipe Opérationnelle d'Hygiène Hospitalière, Marseille (C. Sartor, R. Charrel); Aix Marseille Université, Institut de Recherche et Développement, Microbes Evolution Phylogeny and Infections, Marseille (V. Moal); Assistance Publique Hôpitaux de Marseille, Hôpital Conception, Centre de Néphrologie et Transplantation Rénale, Marseille (V. Moal, J. Solignac); UMR D257 RITMES, Aix-Marseille Université, Assistance Publique Hôpitaux de Marseille, Service de Santé des armées, Marseille (J. Sevestre, S. Ranque, E. Menu); Unité des Virus Émergents, Aix-Marseille Université, Università di Corsica, IRD 190, Inserm 1207, IRBA, Marseille (R. Charrel); Institut Pasteur, Université Paris Cité, National Reference Center for Invasive Mycoses & Antifungals, Mycology Translational Research Group, Paris, France (M. Desnos-Ollivier)
Main Article
Figure 1
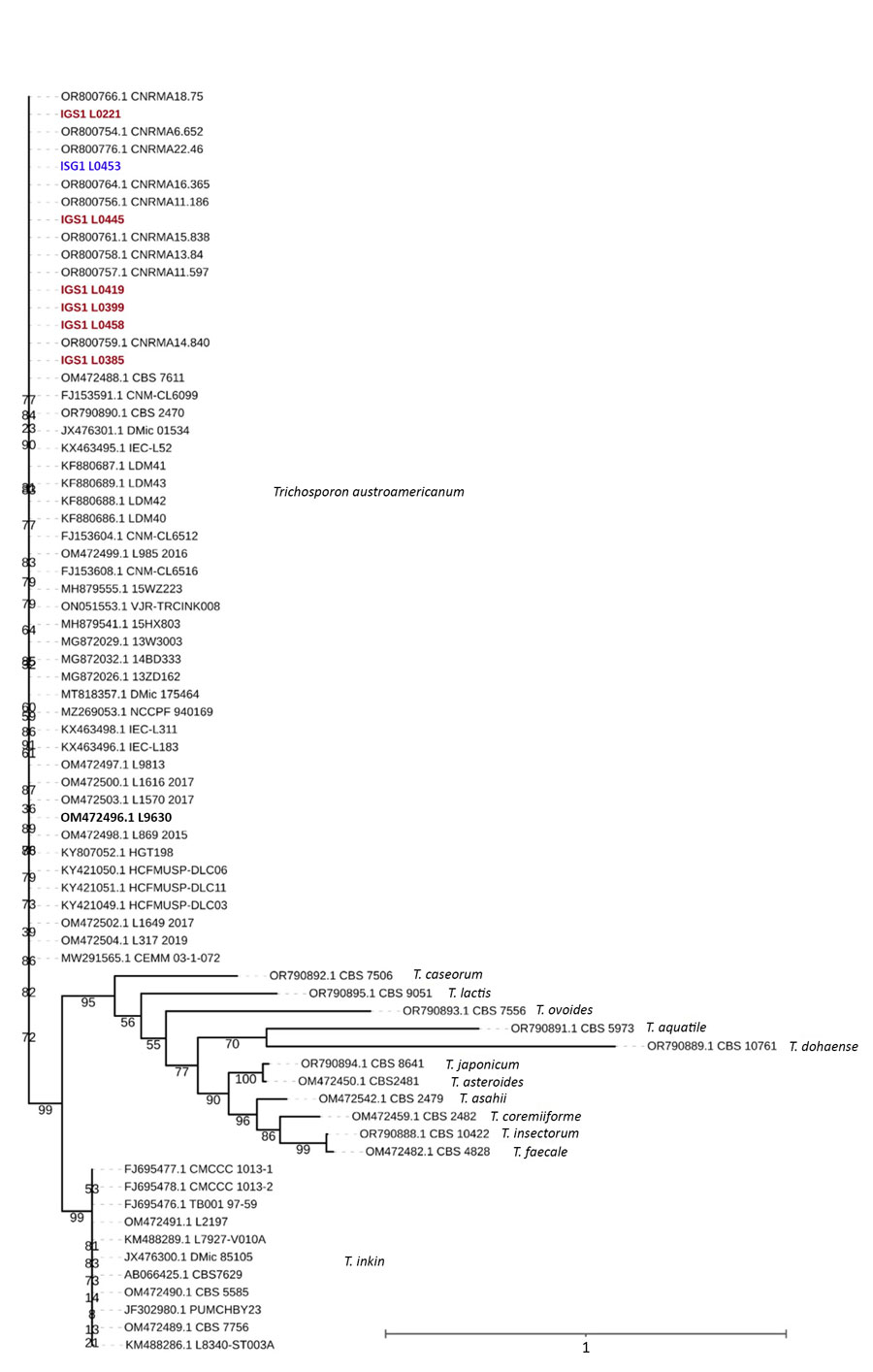
Figure 1. Maximum-likelihood phylogenetic tree of IGS1 sequences from study of Trichosporon austroamericanum infections among hospitalized patients, France, 2022–2024. The tree includes strains isolated from 6 patients (red font) and 1 environmental sample (L0453, blue font), mapped against T. austroamericanum and related species from GenBank (https://www.ncbi.nlm.nih.gov/genbank) and the CBS culture collection (https://wi.knaw.nl/fungal_table). Bold font indicates reference strain CBS 17435. The clustering confirms that the patient and environmental strains belong to T. austroamericanum and form a distinct clade. The tree also shows the relationships between other Trichosporon species, such as T. inkin, T. caseorum, and T. ovoides, and other T. austroamericanum reference strains from the National Reference Center for Invasive Mycoses and Antifungals at Institut Pasteur (https://www.pasteur.fr). Bootstrap values are indicated at the nodes. Scale bar indicates nucleotide substitutions per site. IGS, intergenic spacer region.
Main Article
Page created: September 18, 2025
Page updated: December 10, 2025
Page reviewed: December 10, 2025
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
