Volume 31, Number 11—November 2025
Research Letter
Molecular Evidence of Dengue Virus Serotype 2 in Travelers Returning to Israel from the Sinai Peninsula
Cite This Article
Citation for Media
Abstract
We report 4 dengue cases in travelers returning to Israel from Sharm-El-Sheikh, Egypt, all confirmed as dengue virus type 2 infections. Phylogenetic analysis showed clustering with strains from Pakistan. Our findings provide molecular evidence of dengue circulation in the Sinai desert, highlighting the need for increased awareness among travelers and health authorities.
Dengue virus (DENV) is the most widespread arbovirus globally; its incidence has increased tenfold in the past 2 decades, largely driven by climate change and globalization (1). Although transmission is well documented in Southeast Asia and the Americas, autochthonous emergence is increasingly reported in nonendemic regions, including Europe.
We report 4 confirmed dengue fever cases in travelers returning to Israel after visiting Sharm El-Sheikh, a desert resort city in South Sinai, Egypt, during April–June 2024. Sharm El-Sheikh has not previously been recognized as an area of dengue transmission, the arid environment of the Sinai Peninsula is considered unfavorable for the DENV primary vectors, Aedes mosquitoes.
The cases (Table) were unrelated; travel dates were nonoverlapping and accommodations varied and were located 3–25 km apart. Patients had typical dengue symptoms such as fever, headache, myalgia, and rash. All were hospitalized, received supportive care, and recovered. One patient exhibited meningeal irritation; cerebrospinal fluid testing results were unremarkable, although DENV serotype 2 (DENV-2) RNA was detected by quantitative real-time PCR (cycle threshold 32.5). All samples were collected within 1 week of symptom onset. Serum testing confirmed DENV-2 by multiplex quantitative real-time PCR (2); additional nonstructural protein 1 antigen and IgM/IgG positivity was detected in some cases.
To explore the geographic origin of the DENV-2 cases, we performed DENV whole-genome sequencing. We captured DENV-2 using specific whole-genome primers (https://grubaughlab.com/open-science/amplicon-sequencing); we prepared sequencing libraries using Nextera-XT and ran them on the Illumina NovaSeq (https://www.illumina.com). We generated consensus sequences by mapping to the DENV-2 reference genome (GenBank accession no. NC_001474.2) and deposited resulting sequences into GenBank (Appendix Table). Use of the samples in this study was approved by the Sheba Medical Center Institutional Review Board (approval no. SMC-6190-19).
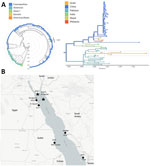
Samples yielded sufficient DENV-2 genome coverage, except in the case of patient no. 4 (possibly because of high cycle threshold [34]), which was excluded. Phylogenetic analysis with global DENV-2 sequences (n = 1,492) clustered the Israel sequences within the Cosmopolitan genotype. All 3 sequences formed a distinct cluster, sharing a common ancestor and differing by 32 mutations from the nearest global strain. The closest related sequences were from Pakistan. The only publicly available sequence geographically close to Sinai, from the United Arab Emirates in 2023, clustered separately within another Cosmopolitan lineage with strains from China, India, and Bangladesh (Figure, panel A).
Our findings describe 4 confirmed DENV-2 infections in travelers from Sharm El-Sheikh, Egypt, a city in the arid Sinai Peninsula, previously considered unsuitable for Aedes mosquitoes and without previous dengue reports. Genomic analysis showed clustering of cases, likely from a single outbreak, most closely related to strains from Pakistan. Aside from 1 United Arab Emirates 2023 sequence clustering separately, no recent data from Sinai are available, underscoring a major surveillance gap. Those results align with reports of DENV-2 spread along the Red Sea and recent cases in Florence, Italy (8).
During the past 2 decades, Ae. aegypti mosquito populations have expanded in Egypt, especially along the Red Sea coast (Figure, panel B), correlating with dengue outbreaks. However, no entomologic data exist for Sinai. The arid climate challenges mosquito survival, but clustering of cases in 1 resort area suggests local adaptation, possibly supported by urban microhabitats (9). Maritime and air travel might drive repeated introductions of Ae. aegypti mosquitos and DENV into the Red Sea region. However, the pattern of DENV-2 outbreaks in Red Sea port cities support maritime transport as a key driver of spread (6,7,10). The daily ferries from Hurghada, where dengue recently emerged, to Sharm El-Sheikh might be especially relevant (Figure, panel B). Genetic data from a 2019 Jizan outbreak and strains from Saudi Arabia (1992–2014) further suggest multiple introductions linked to an imported DENV-2 variant genetically similar to strains from Malaysia, Singapore, Korea, and China (6). Additional analyses from the DENV-2 strains isolated in Saudi Arabia during 1992–2014 reveal strong clustering with viruses from countries that contribute the largest numbers of Hajj and Umrah pilgrims: Indonesia, Pakistan, and India (10). Indeed, phylogenetic analysis shows that our dengue sequences are closest to recent strains from Pakistan. However, the scarcity of sequences from Egypt and neighboring regions limits inference on viral origin, circulation, and distribution, and observed variability suggests undersampling and additional undetected cases.
This report of 4 cases over 3 months in different localities of Sharm El-Sheikh suggests sustained DENV-2 transmission and emphasizes the importance of enhanced vector surveillance and control, providing an alert to public health authorities. The genetic data presented might help address gaps in regional DENV sequence reporting and contribute to understanding its molecular epidemiology and origins.
Dr. Zuckerman leads the Bioinformatics and Genomics Center at Israel’s Central Virology Laboratory, Ministry of Health, and is affiliated with Tel Aviv University’s School of Public Health. Her work focuses on genomic surveillance, molecular epidemiology, and bioinformatics applications in the study of viral pathogens
References
- World Health Organization. Dengue and severe dengue [2025 Mar 1]. https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue.
- Santiago GA, Vergne E, Quiles Y, Cosme J, Vazquez J, Medina JF, et al. Analytical and clinical performance of the CDC real time RT-PCR assay for detection and typing of dengue virus. PLoS Negl Trop Dis. 2013;7:
e2311 . DOIPubMedGoogle Scholar - Kholedi AAN, Balubaid O, Milaat W, Kabbash IA, Ibrahim A. Factors associated with the spread of dengue fever in Jeddah Governorate, Saudi Arabia. East Mediterr Health J. 2012;18:15–23. DOIPubMedGoogle Scholar
- El-Kady AM, Osman HA, Alemam MF, Marghani D, Shanawaz MA, Wakid MH, et al. Circulation of dengue virus serotype 2 in humans and mosquitoes during an outbreak in El Quseir City, Egypt. Infect Drug Resist. 2022;15:2713–21. DOIPubMedGoogle Scholar
- Desogi M, Ali M, Gindeel N, Khalid F, Abdelraheem M, Alnaby A, et al. Detection of dengue virus serotype 4 in Sudan. East Mediterr Health J. 2023;29:436–41. DOIPubMedGoogle Scholar
- Dafalla O, Abdulhaq AA, Almutairi H, Noureldin E, Ghzwani J, Mashi O, et al. The emergence of an imported variant of dengue virus serotype 2 in the Jazan region, southwestern Saudi Arabia. Trop Dis Travel Med Vaccines. 2023;9:5. DOIPubMedGoogle Scholar
- Frank C, Lachmann R, Wilking H, Stark K. Increase in dengue fever in travellers returning from Egypt, Germany 2023. Euro Surveill. 2024;29:
2400042 . DOIPubMedGoogle Scholar - Manciulli T, Zammarchi L, Lagi F, Fiorelli C, Mencarini J, Fognani M, et al. Emergence of dengue fever: sentinel travellers uncover outbreak in Sharm El-Sheikh, Egypt, May 2024. J Travel Med. 2024;31:taae080. DOIGoogle Scholar
- Newman EA, Feng X, Onland JD, Walker KR, Young S, Smith K, et al. Defining the roles of local precipitation and anthropogenic water sources in driving the abundance of Aedes aegypti, an emerging disease vector in urban, arid landscapes. Sci Rep. 2024;14:2058. DOIPubMedGoogle Scholar
- El-Kafrawy SA, Sohrab SS, Ela SA, Abd-Alla AM, Alhabbab R, Farraj SA, et al. Multiple introductions of dengue 2 virus strains into Saudi Arabia from 1992 to 2014. Vector Borne Zoonotic Dis. 2016;16:391–9. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleOriginal Publication Date: December 04, 2025
1These authors contributed equally to this article.
Table of Contents – Volume 31, Number 11—November 2025
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Neta S. Zuckerman, Central Virology Laboratory, Sheba Medical Center, 2 Derech Sheba, Ramat-Gan, Israel
Top