Volume 31, Number 11—November 2025
Research Letter
Molecular Evidence of Dengue Virus Serotype 2 in Travelers Returning to Israel from the Sinai Peninsula
Figure
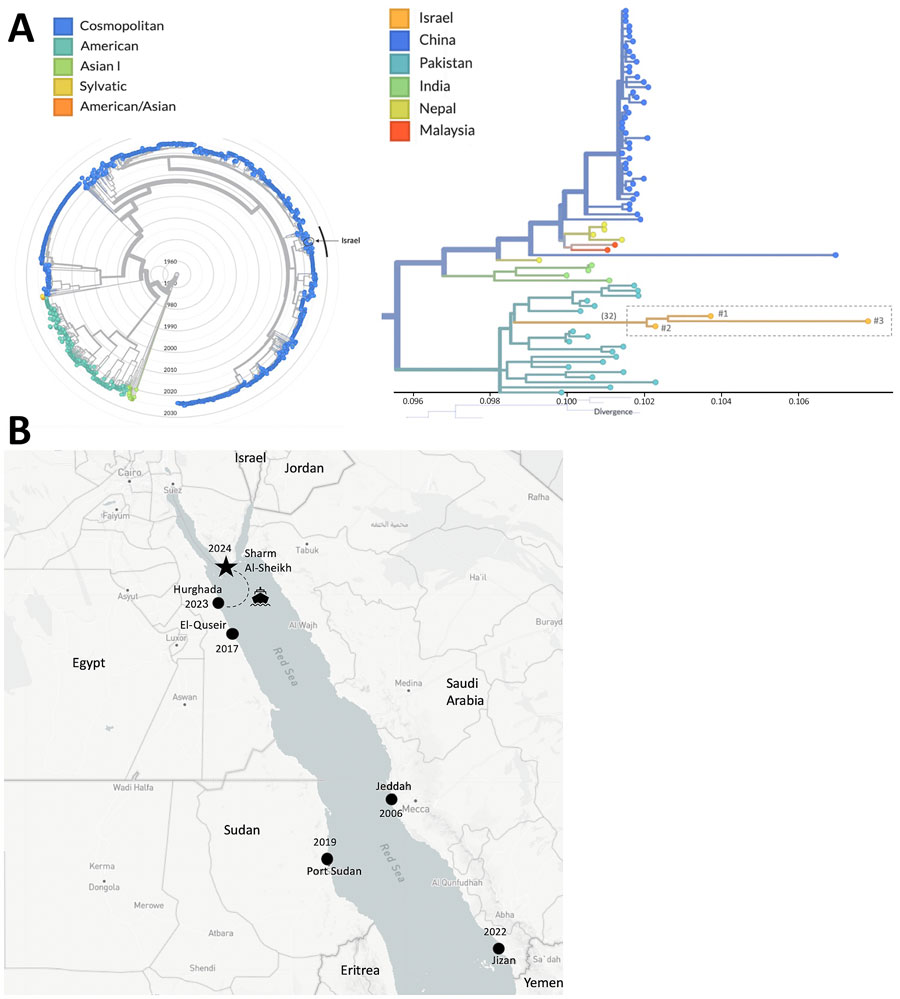
Figure. Epidemiology, phylogeny, and geography of dengue virus (DENV) serotype 2 (DENV-2) circulation in the Red Sea region in study of molecular evidence of DENV-2 emergence from travelers returning to Israel from the Sinai Peninsula. A) Phylogenetic analysis of DENV-2 whole-genome sequences. Phylogenetic tree was constructed using the samples from Israel sequenced in this study (n = 3) alongside global DENV-2 sequences from the past decade (n = 1,492) available in the DENV-2 Nextstrain build (https://nextstrain.org/dengue/denv2/genome). The radial tree illustrates the global DENV-2 genotypes, highlighting the Israel sequences (n = 3, circled) within the broader dataset. Nodes are color-coded by DENV genotype. The cluster marked by a black arc is magnified in the right portion. The rectangular tree focuses on the cluster containing the Israeli sequences (n = 3), showing the genetic divergence within the group. Nodes are color-coded by the country of origin, and the number of mutations connecting the Israeli cluster to the nearest ancestor is indicated in parentheses. B) Dengue fever outbreak locations, including the current outbreak in Sharm El-Sheikh (star) and outbreaks reported in the region in recent years (black circles labeled by year): Jeddah 2006 (3), El-Quseir 2017 (4), Port Sudan 2019 (5), Jizan 2022 (6), and Hurghada 2023 (7).
References
- World Health Organization. Dengue and severe dengue [2025 Mar 1]. https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue.
- Santiago GA, Vergne E, Quiles Y, Cosme J, Vazquez J, Medina JF, et al. Analytical and clinical performance of the CDC real time RT-PCR assay for detection and typing of dengue virus. PLoS Negl Trop Dis. 2013;7:
e2311 . DOIPubMedGoogle Scholar - Kholedi AAN, Balubaid O, Milaat W, Kabbash IA, Ibrahim A. Factors associated with the spread of dengue fever in Jeddah Governorate, Saudi Arabia. East Mediterr Health J. 2012;18:15–23. DOIPubMedGoogle Scholar
- El-Kady AM, Osman HA, Alemam MF, Marghani D, Shanawaz MA, Wakid MH, et al. Circulation of dengue virus serotype 2 in humans and mosquitoes during an outbreak in El Quseir City, Egypt. Infect Drug Resist. 2022;15:2713–21. DOIPubMedGoogle Scholar
- Desogi M, Ali M, Gindeel N, Khalid F, Abdelraheem M, Alnaby A, et al. Detection of dengue virus serotype 4 in Sudan. East Mediterr Health J. 2023;29:436–41. DOIPubMedGoogle Scholar
- Dafalla O, Abdulhaq AA, Almutairi H, Noureldin E, Ghzwani J, Mashi O, et al. The emergence of an imported variant of dengue virus serotype 2 in the Jazan region, southwestern Saudi Arabia. Trop Dis Travel Med Vaccines. 2023;9:5. DOIPubMedGoogle Scholar
- Frank C, Lachmann R, Wilking H, Stark K. Increase in dengue fever in travellers returning from Egypt, Germany 2023. Euro Surveill. 2024;29:
2400042 . DOIPubMedGoogle Scholar - Manciulli T, Zammarchi L, Lagi F, Fiorelli C, Mencarini J, Fognani M, et al. Emergence of dengue fever: sentinel travellers uncover outbreak in Sharm El-Sheikh, Egypt, May 2024. J Travel Med. 2024;31:taae080. DOIGoogle Scholar
- Newman EA, Feng X, Onland JD, Walker KR, Young S, Smith K, et al. Defining the roles of local precipitation and anthropogenic water sources in driving the abundance of Aedes aegypti, an emerging disease vector in urban, arid landscapes. Sci Rep. 2024;14:2058. DOIPubMedGoogle Scholar
- El-Kafrawy SA, Sohrab SS, Ela SA, Abd-Alla AM, Alhabbab R, Farraj SA, et al. Multiple introductions of dengue 2 virus strains into Saudi Arabia from 1992 to 2014. Vector Borne Zoonotic Dis. 2016;16:391–9. DOIPubMedGoogle Scholar
1These authors contributed equally to this article.