Volume 31, Number 12—December 2025
Dispatch
Macrolide Resistance and P1 Cytadhesin Genotyping of Mycoplasma pneumoniae during Outbreak, Canada, 2024–2025
Figure 2
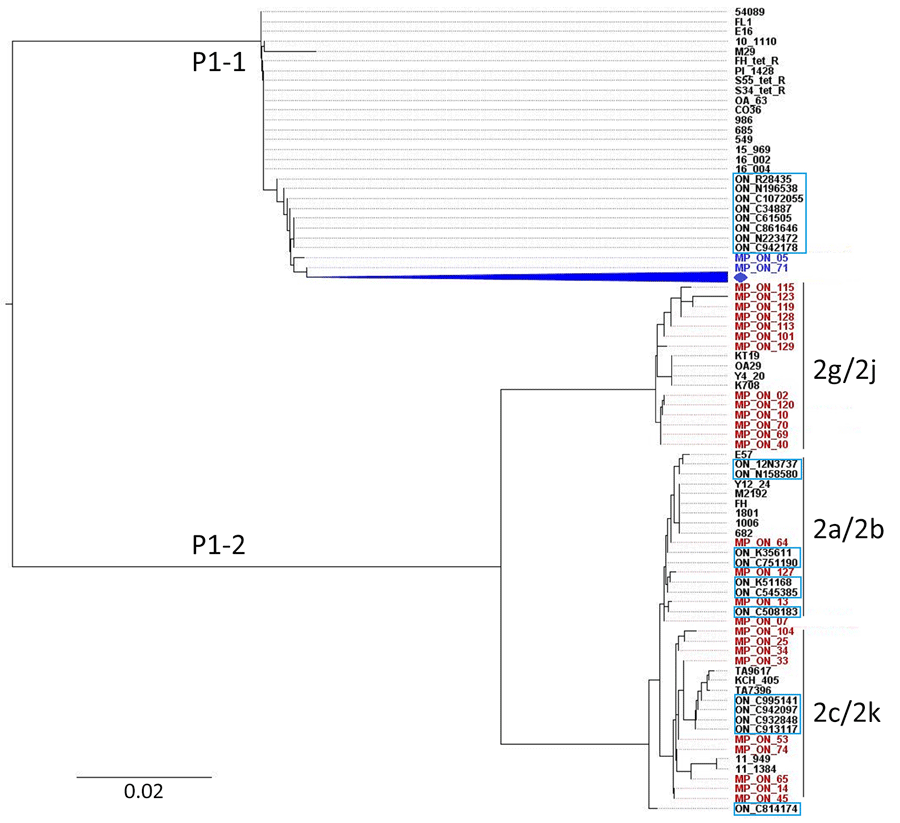
Figure 2. Phylogenetic analysis of Mycoplasma pneumoniae based on P1 cytadhesin–RepMP4 genotyping during 2024–2025 outbreak, Hamilton, Ontario, Canada. An unrooted tree was constructed using the neighbor-joining method with the Tamura-Nei model in MEGA X (https://www.megasoftware.net) using aligned sequences generated using Clustal Omega (https://www.ebi.ac.uk/jdispatcher/msa/clustalo). Strains highlighted in blue and red represent the P1-1 and P1-2 type strains assessed in this study. Strains in the light blue boxes indicate previously reported strains from Ontario during 2011–2012 (8). Strains shown in black represent reference RepMP4 sequences from M. pneumoniae obtained in other countries, representing P1 types and variants (Appendix Table 3). Blue diamond indicates remaining 104 P1-1 strains from this study.
References
- Waites KB, Xiao L, Liu Y, Balish MF, Atkinson TP. Mycoplasma pneumoniae from the respiratory tract and beyond. Clin Microbiol Rev. 2017;30:747–809. DOIPubMedGoogle Scholar
- Chen YC, Hsu WY, Chang TH. Macrolide-resistant Mycoplasma pneumoniae infections in pediatric community-acquired pneumonia. Emerg Infect Dis. 2020;26:1382–91. DOIPubMedGoogle Scholar
- Upadhyay P, Singh V. Mycoplasma pneumoniae outbreak in 2023: post-pandemic resurgence of an atypical bacterial pathogen. Cureus. 2024;16:
e58757 . DOIPubMedGoogle Scholar - Meyer Sauteur PM, Beeton ML; European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group for Mycoplasma and Chlamydia Infections (ESGMAC), and the ESGMAC Mycoplasma pneumoniae Surveillance (MAPS) study group. Mycoplasma pneumoniae: delayed re-emergence after COVID-19 pandemic restrictions. Lancet Microbe. 2024;5:e100–1. DOIPubMedGoogle Scholar
- Edouard S, Boughammoura H, Colson P, La Scola B, Fournier PE, Fenollar F. Large-scale outbreak of Mycoplasma pneumoniae infection, Marseille, France, 2023–2024. Emerg Infect Dis. 2024;30:1481–4. DOIPubMedGoogle Scholar
- Public Health Ontario. Mycoplasma pneumoniae testing at Public Health Ontario [cited 2025 Jun 11]. https://www.publichealthontario.ca/-/media/Documents/M/24/mycoplasma-pneumoniae-testing-pho.pdf
- Jiang FC, Wang RF, Chen P, Dong LY, Wang X, Song Q, et al. Genotype and mutation patterns of macrolide resistance genes of Mycoplasma pneumoniae from children with pneumonia in Qingdao, China, in 2019. J Glob Antimicrob Resist. 2021;27:273–8. DOIPubMedGoogle Scholar
- Eshaghi A, Memari N, Tang P, Olsha R, Farrell DJ, Low DE, et al. Macrolide-resistant Mycoplasma pneumoniae in humans, Ontario, Canada, 2010-2011. Emerg Infect Dis. 2013;19:1525–7. DOIPubMedGoogle Scholar
1These first authors contributed equally to this article.