Volume 31, Number 12—December 2025
Dispatch
Human Infection by Zoonotic Eye Fluke Philophthalmus lacrymosus, South America
Figure 2
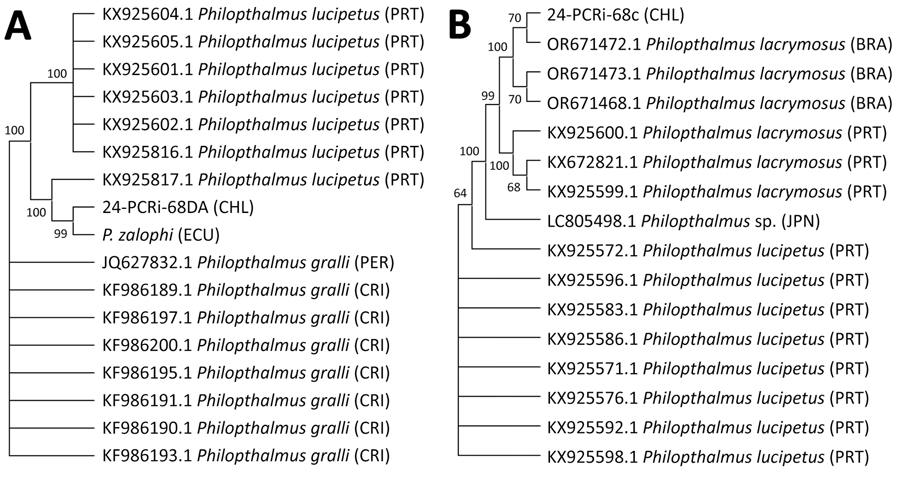
Figure 2. Maximum-likelihood phylogenetic tree constructed from 17 internal transcribed spacer 2 sequences (trimmed alignment 673 bp) (A) and 16 cytochrome c oxidase I sequences (trimmed alignment 365 bp) (B) of Philophthalmus lacrymosus fluke extracted from conjunctiva of a female traveler from England in Chile. Consensus trees were inferred from 1,000 replicates by using the Kimura 2-parameter test in MEGA 11 (https://www.megasoftware.net). Bootstrap values at the nodes indicate the percentages of replicates in which the sequences clustered together. Sequence codes include GenBank accession numbers and parasite information. In parentheses, letter codes indicate the country of origin. Sequences from this case report are 24-PCRi-68DA_CHL (GenBank accession no. PX240011) and 24-PCRi-68c_CHL (accession no. PX238763). BRA, Brazil; CHL, Chile; CRI, Costa Rica; ECU, Ecuador; JPN, Japan; PER, Peru; PRT, Portugal.
1These authors contributed equally to this article.