Volume 31, Number 12—December 2025
Research Letter
Carbapenem-Resistant Salmonella Typhi Infection in Traveler Returning to Germany from India, 2024
Cite This Article
Citation for Media
Abstract
We report on a carbapenem-, extended spectrum β-lactam-, fluoroquinolone-, and tetracycline-resistant Salmonella enterica serovar Typhi strain in a patient returning to Germany from India. Considering the recent emergence of extensively drug-resistant Salmonella Typhi strains, further expansion of antibiotic resistance to carbapenems poses a serious threat for typhoid fever treatment.
Extensively drug-resistant (XDR) Salmonella enterica serovar Typhi, belonging to the H58 haplotype, was first identified in Sindh, Pakistan, in 2016 (1). Since then, those strains have been reported worldwide, mainly in association with travel to Pakistan. XDR Salmonella Typhi exhibit a multidrug-resistant (MDR) phenotype, including resistance to chloramphenicol, ampicillin, and sulfamethoxazole/trimethoprim, along with additional resistance to fluoroquinolones and third-generation cephalosporins. Consequently, therapeutic options for treating infections caused by XDR strains are primarily limited to the macrolide azithromycin and carbapenems. Salmonella Typhi strains resistant to carbapenems, azithromycin, or both have been reported occasionally. Carbapenem-resistant strains isolated in Pakistan harbored genes encoding VIM, GES, or NDM-5 carbapenemases (2,3). The respective NDM-5–positive strain showed the XDR phenotype and was phylogenetically assigned to the H58 haplotype. Recently, another case study described a non–XDR NDM-5–producing Salmonella Typhi isolate from India, which revealed additional resistance to fluoroquinolones and third-generation cephalosporins but remained susceptible to chloramphenicol, sulfamethoxazole/trimethoprim, and azithromycin (4).
We report an NDM-5–producing Salmonella Typhi strain isolated from a patient from Germany after returning from India. The patient, an experienced traveler to India who was last vaccinated with the typhoid polysaccharide vaccine in June 2021, undertook a 4-week round trip through several states in southwest India in September and October 2024. Upon return to Germany, the patient had onset of mild gastrointestinal symptoms, including diarrhea and abdominal pain. Symptoms gradually worsened over 2 weeks, prompting the patient to seek outpatient medical attention at a medical practice and a clinic, where stool and blood samples were collected. Blood tests were suggestive of a bacterial infection, and an empiric 3-day course of ciprofloxacin was commenced. However, after completion of the antibiotic therapy, the patient’s condition further deteriorated. Specifically, the patient had onset of fever (38.5°C) and severe headaches. Molecular stool diagnostics provided positive PCR signals for Salmonella and Shigella spp., but stool culture only resulted in growth of Salmonella spp. We subtyped the retrieved isolate (no. 24-09143) as Salmonella Typhi; phenotypic antimicrobial susceptibility testing according to European Committee on Antimicrobial Susceptibility Testing guidelines determined resistance to fluoroquinolones, tetracyclines, and β-lactam antibiotics, including penicillins, third-generation cephalosporins, and carbapenems (Table). The strain was susceptible to chloramphenicol, sulfamethoxazole/trimethoprim, and azithromycin, distinguishing it from MDR or XDR Salmonella Typhi and making appropriate treatment possible. The patient’s illness was successfully treated with a 14-day course of oral sulfamethoxazole/trimethoprim, and the patient made a full clinical recovery without requiring hospitalization. Three follow-up stool samples remained culture-negative.
We performed whole-genome analysis by using Illumina (https://www.illumina.com) (European Nucleotide Archive [ENA] accession no. ERR15390137) and Oxford Nanopore (https://nanoporetech.com) (ENA accession no. ERR15647836) technologies. We compared resistance determinants of strain 24-09143 from the reported case with published Salmonella Typhi strains from Pakistan showing XDR, azithromycin, and carbapenem resistance (3), and from India, characterized by either extended-spectrum β-lactamase (ESBL) (5) or NDM-5 production (4) (Table). Resistance determinants of strain 24-09143 included tet(A) for tetracycline, qnrS1 for quinolone, blaCTX-M-15 for cephalosporin, and blaNDM-5 for carbapenem. On the basis of the hybrid assembly of the long and short read data (obtained by using unicycler version 0.4.8, https://github.com/rrwick/Unicycler), we located the resistance genes on 2 putative conjugative plasmids of replicon types IncFIB (qnrS1 and tet(A), blaCTX-M-15, 73.3 kb) and IncX3 (blaNDM-5, 46.2 kb). blaCTX-M-15 is regularly found in ESBL-producing Salmonella Typhi of the Pakistan XDR sublineage, either plasmidborne or integrated in the genome (1,6), and has also been described in non–XDR Salmonella Typhi from India (5,7). Although carbapenemase production has thus far been observed rarely in Salmonella Typhi (3,4), IncX3 plasmids carrying blaNDM-5 have been reported in nontyphoidal Salmonella (8,9) and in other enterobacterial genera.
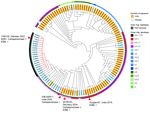
Phylogenetic analysis assigned the strain to the H58 haplotype but distinguished it from the previously described NDM-5–positive XDR Salmonella Typhi from Pakistan (strain no. 1790125; ENA accession no. SRR22801766) (3) by 22 single-nucleotide polymorphisms (SNPs) (Figure). However, the analysis revealed a very close relationship to the recently described NDM-5–positive isolate from India (0 SNPs; strain no. IOB-SWH-1; ENA accession no. SRR32461882) (4) and to a representative of a carbapenemase-negative ESBL-producing S. Typhi lineage also orginating from India (5) (4 SNPs; strain Gurgaon01; ENA accession no. ERR3527963) (Figure). The presence of the blaNDM-5 gene in representatives of different Salmonella Typhi sublineages presumably indicates independent acquisitions in the XDR sublineage from Pakistan and an ESBL-producing sublineage from India, which might increase the risk for global dissemination of carbapenem resistance in which transmission is enabled by travel and migration. This risk is exemplified for the reported case, in which an individual contracted the NDM-5–producing Salmonella Typhi while traveling in India and brought the strain to Germany upon return. Alarmingly, the increasing occurrence of XDR, ESBL-producing, and carbapenemase-producing strains severely limits effective treatment options for typhoid fever. This finding emphasizes the need for comprehensive antimicrobial susceptibility testing of clinical Salmonella Typhi isolates to ensure appropriate treatment, while avoiding the use of last-line antibiotics when they are not necessary. It also highlights the importance of prevention measures, such as improved sanitation, access to clean water, and typhoid vaccination (or revaccination).
Dr. Simon works as scientist for the Department of Infectious Diseases at the Robert Koch Institute and Germany’s National Reference Center for Salmonella and Other Bacterial Enteric Pathogens. Her research interests include integrated genomic surveillance, antimicrobial resistance, and risk profiling of clinical Salmonella strains.
Acknowledgment
We thank the National Reference Center for Salmonella and Other Bacterial Enteric Pathogens laboratory assistants for their excellent technical support. We also thank the Sequencing Core Facility of the Genome Competence Centre, Robert Koch Institute, for providing high-quality whole genome data for the Salmonella Typhi strains from Germany.
References
- Klemm EJ, Shakoor S, Page AJ, Qamar FN, Judge K, Saeed DK, et al. Emergence of an extensively drug-resistant Salmonella enterica serovar Typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. mBio. 2018;9:e00105–18.PubMedGoogle Scholar
- Ain Q, Tahir M, Sadaqat A, Ayub A, Awan AB, Wajid M, et al. First detection of extensively drug-resistant Salmonella Typhi isolates harboring VIM and GES genes for carbapenem resistance from Faisalabad, Pakistan. Microb Drug Resist. 2022;28:1087–98.PubMedGoogle Scholar
- Nizamuddin S, Khan EA, Chattaway MA, Godbole G. Case of carbapenem-resistant Salmonella Typhi infection, Pakistan, 2022. Emerg Infect Dis. 2023;29:2395–7.PubMedGoogle Scholar
- Vasanthaiah S, Takey P, Selvam PK, Mohan S, Kiran R, Roohi S, et al. Genomic perspectives on NDM Salmonella Typhi, and a case report from India. Infection. 2025;53:2053–9.PubMedGoogle Scholar
- Sah R, Donovan S, Seth-Smith HMB, Bloemberg G, Wüthrich D, Stephan R, et al. A novel lineage of ceftriaxone-resistant Salmonella Typhi from India that is closely related to XDR S. Typhi found in Pakistan. Clin Infect Dis. 2020;71:1327–30.PubMedGoogle Scholar
- Nair S, Chattaway M, Langridge GC, Gentle A, Day M, Ainsworth EV, et al. ESBL-producing strains isolated from imported cases of enteric fever in England and Wales reveal multiple chromosomal integrations of blaCTX-M-15 in XDR Salmonella Typhi. J Antimicrob Chemother. 2021;76:1459–66.PubMedGoogle Scholar
- Thirumoorthy TP, Jacob JJ, Velmurugan A, Teekaraman MP, Shah B, Iyer V, et al. Recent emergence of cephalosporin-resistant Salmonella Typhi in India due to the endemic clone acquiring IncFIB(K) plasmid encoding blaCTX-M-15 gene. Microbiol Spectr. 2025;13:
e0087524 .PubMedGoogle Scholar - Ramírez-Castillo FY, Guerrero-Barrera AL, Avelar-González FJ. An overview of carbapenem-resistant organisms from food-producing animals, seafood, aquaculture, companion animals, and wildlife. Front Vet Sci. 2023;10:
1158588 .PubMedGoogle Scholar - Wu Y, Jiang T, Bao D, Yue M, Jia H, Wu J, et al. Global population structure and genomic surveillance framework of carbapenem-resistant Salmonella enterica. Drug Resist Updat. 2023;68:
100953 .PubMedGoogle Scholar - Wong VK, Baker S, Connor TR, Pickard D, Page AJ, Dave J, et al.; International Typhoid Consortium. An extended genotyping framework for Salmonella enterica serovar Typhi, the cause of human typhoid. Nat Commun. 2016;7:12827.PubMedGoogle Scholar
Figure
Table
Cite This ArticleOriginal Publication Date: December 23, 2025
Table of Contents – Volume 31, Number 12—December 2025
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Antje Flieger, Robert Koch-Institut, Unit Enteropathogenic Bacteria and Legionella, National Reference Center for Salmonella and other Bacterial other Bacterial Enteric Pathogens, Burgstr. 37, D-38855 Wernigerode, Germany
Top