Volume 31, Number 2—February 2025
CME ACTIVITY - Research
Streptococcus pyogenes emm Type 3.93 Emergence, the Netherlands and England
Figure 3
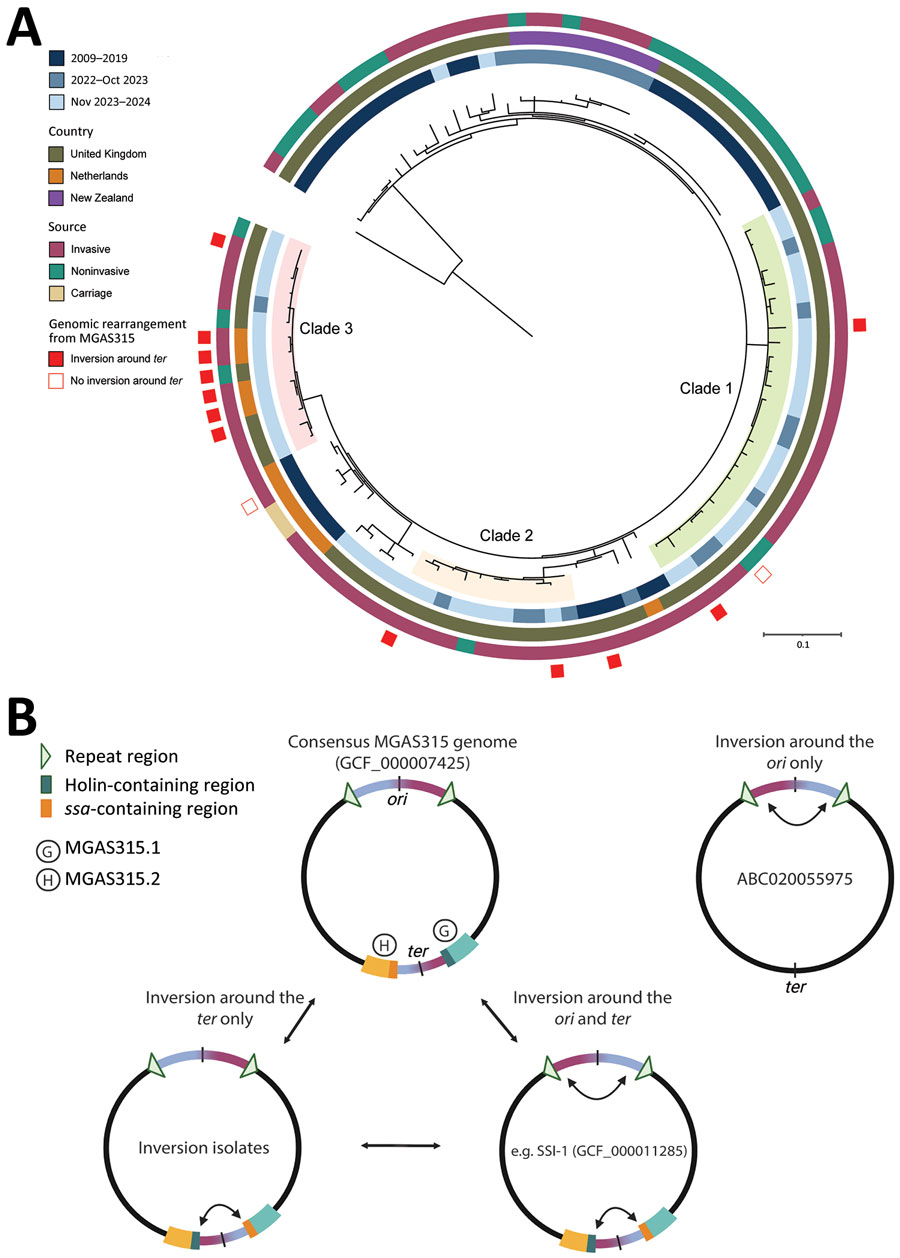
Figure 3. Genetic information about emm3.93 Streptococcus pyogenes isolates collected in the Netherlands and England, February 1, 2023–March 31, 2024, and reference isolates. A) Maximum-likelihood phylogenetic tree of 114 parsimony informative sites and 255 distinct site patterns from the core genome single-nucleotide polymorphism alignment of 104 emm3.93 isolates from the Netherlands, England, and New Zealand (2009–2024), compared with MGAS315 reference genome (RefSeq accession no. GCF_000007425). The tree was rooted on MGAS315, and colored rings outside the tree represent year of isolation, country, source, and presence of genomic inversion around the ter. Scale bar represents substitutions per site. B) Schematic of the 4 genomic conformations detected in the emm3 lineage. When compared with the consensus reference genome of strain MGAS315, an inversion around both the ori and ter, or around the ter alone (as detected in 11/12 surge isolates) is observed. Genome ABC020055975 indicates a strain lacking prophages MGAS315.1 and MGAS315.2 but with a detected inversion around the ori. G, MGAS315.1; H, MGAS315.2.
1These authors contributed equally.