Volume 31, Number 2—February 2025
Research
Respiratory Shedding of Infectious SARS-CoV-2 Omicron XBB.1.41.1 Lineage among Captive White-Tailed Deer, Texas, USA
Figure 1
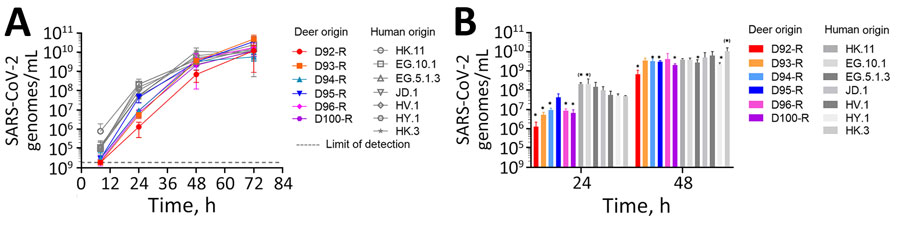
Figure 1. Multistep growth characteristics of contemporaneous strains isolated from captive white-tailed deer and humans in study of respiratory shedding of infectious SARS-CoV-2 Omicron XBB 1.41.1 lineage among captive white-tailed deer, Texas, USA, November 2023. A) Vero E6-TMPRSS2-T2A-ACE2 cells were inoculated with SARS-CoV-2 recovered from deer (D92-R to D100-R) or from human clinical nasopharyngeal samples (EG.10.1, EG.5.1.3, HK.11, JD.1, HV.1, HY.1, HK.3) at a multiplicity of infection of 0.002. Samples of the supernatant were collected and titrated by 1-step quantitative PCR. Lower limit of detection is indicated. B) Averaged data for the 24-hour and 48-hour timepoints. Statistically significant differences after 1-way analysis of variance with Tukey correction are indicated by asterisks for values significantly lower than aggregated human samples and asterisks in parentheses for values significantly higher than aggregated human samples. Error bars indicate SDs calculated from 3 replicates. We added “-R” to the name of each animal identification number to indicate that samples used in these experiments were recovered from the initial virus isolation step.