Volume 31, Number 2—February 2025
Research
Respiratory Shedding of Infectious SARS-CoV-2 Omicron XBB.1.41.1 Lineage among Captive White-Tailed Deer, Texas, USA
Figure 2
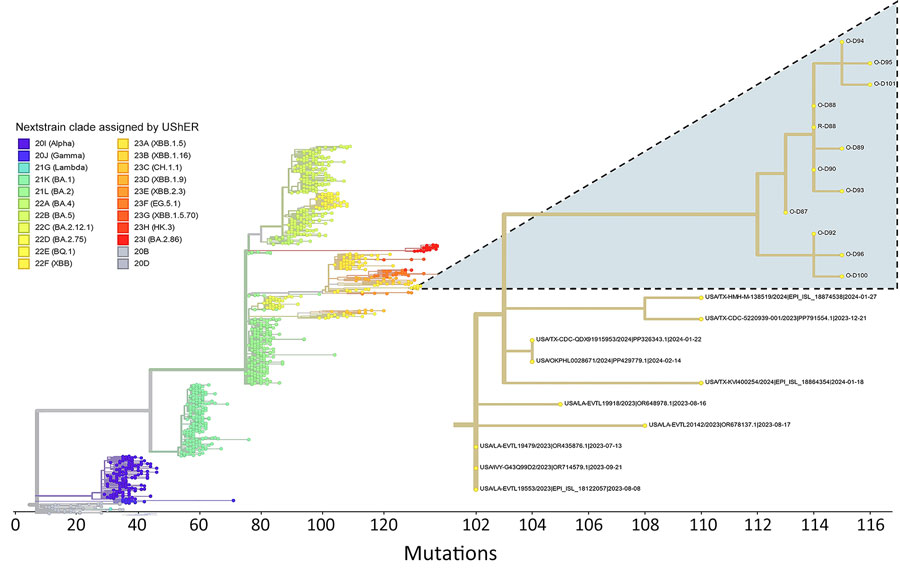
Figure 2. Phylogenetic context of SARS-CoV-2 from captive white-tailed deer in study of respiratory shedding of infectious SARS-CoV-2 Omicron XBB 1.41.1 lineage, Texas, USA, November 2023. Genome sequences from deer were compared in the main phylogenetic tree with genomes representative of the main virus Nexstrain clades as assigned by UShER (https://genome.ucsc.edu/cgi-bin/hgPhyloPlace) (28). The secondary tree at right details the 22F (XBB) clade and displays the placement of genomes obtained in our study (highlighted clade; O indicates oral/nasal swab, R indicates rectal swab) in relation to 10 of the most closely related samples deposited in GISAID (https://www.gisaid.org) as of August 14, 2024.
Page created: December 30, 2024
Page updated: January 31, 2025
Page reviewed: January 31, 2025
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.