Volume 31, Number 2—February 2025
Dispatch
Diphtheria Toxin–Producing Corynebacterium ramonii in Inner-City Population, Vancouver, British Columbia, Canada, 2019–2023
Figure
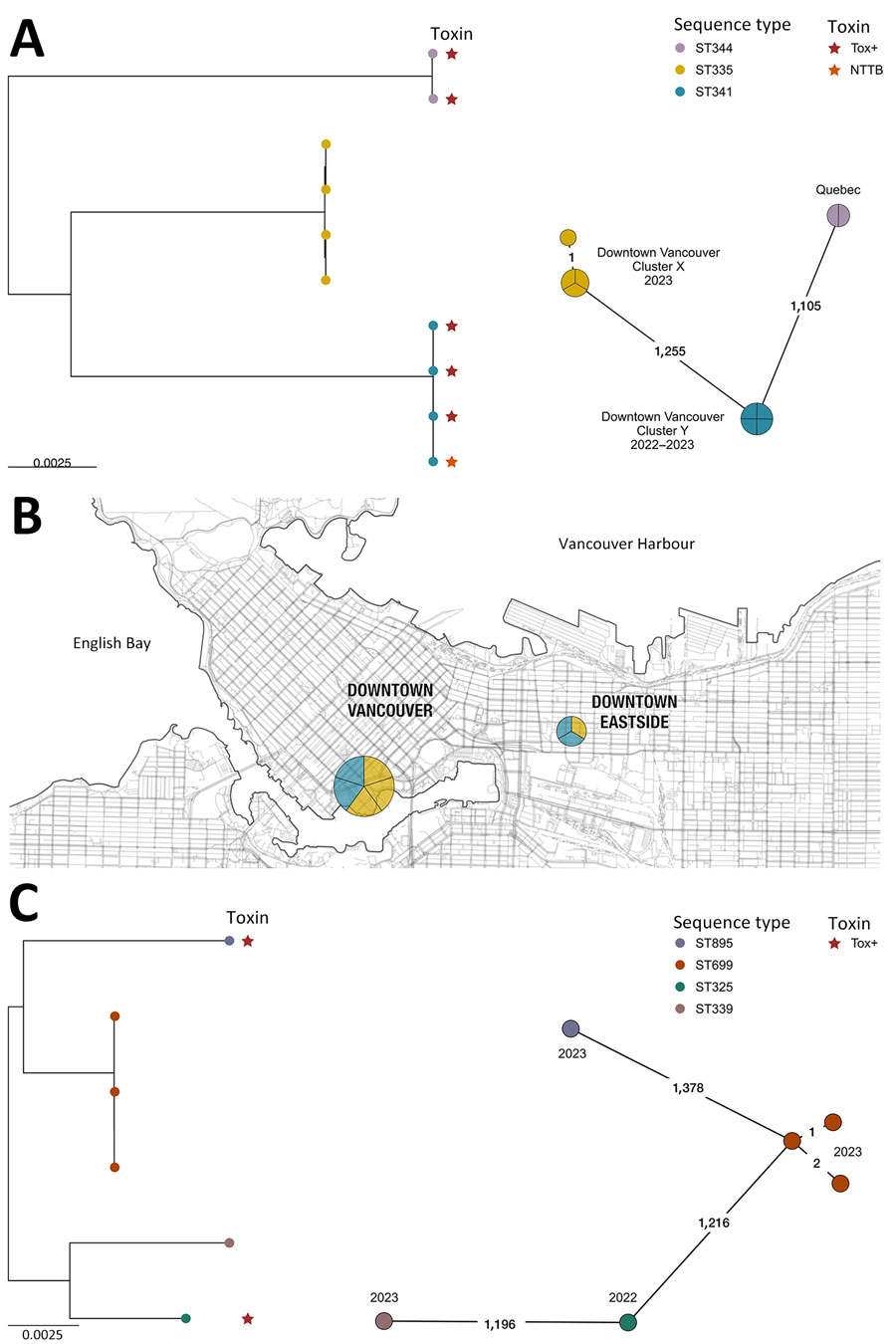
Figure. Phylogenetic analyses and location of Corynebacterium ramonii and C. ulcerans isolates in study of diphtheria toxin–producing C. ramonii in inner-city population, Vancouver, British Columbia, Canada, 2019–2023. Trees were rooted at midpoint. Colored circles indicate sequence type and colored stars indicate diphtheria toxin status. Numbers on lines in the spanning trees indicate number of allelic differences between sequence types; years the specimens were collected are indicated at spanning tree nodes. A) Maximum-likelihood phylogeny (left side) and minimum spanning tree (right side) of 8 C. ramonii isolates from this study (British Columbia) together with 2 isolates, LSPQ-04227 and LSPQ-04228, from Quebec, Canada (1); 2 clusters of infections can be observed, corresponding to ST335 and ST341 isolates. B) Spatial map of C. ramonii isolates from this study, all originating from downtown Vancouver. C) Maximum-likelihood phylogeny (left side) and minimum spanning tree (right side) of 6 C. ulcerans isolates from this study (British Columbia). Scale bars indicates nucleotide substitutions per site. NTTB, nontoxigenic tox gene–bearing; ST, sequence type; Tox+, tox gene and toxin both present.
References
- Crestani C, Arcari G, Landier A, Passet V, Garnier D, Brémont S, et al. Corynebacterium ramonii sp. nov., a novel toxigenic member of the Corynebacterium diphtheriae species complex. Res Microbiol. 2023;174:
104113 . DOIPubMedGoogle Scholar - Otshudiema JO, Acosta AM, Cassiday PK, Hadler SC, Hariri S, Tiwari TSP. Respiratory illness caused by Corynebacterium diphtheriae and C. ulcerans, and use of diphtheria antitoxin in the United States, 1996–2018. Clin Infect Dis. 2021;73:e2799–806. DOIPubMedGoogle Scholar
- Moore LSP, Leslie A, Meltzer M, Sandison A, Efstratiou A, Sriskandan S. Corynebacterium ulcerans cutaneous diphtheria. Lancet Infect Dis. 2015;15:1100–7. DOIPubMedGoogle Scholar
- Bernard KA, Pacheco AL, Burdz T, Wiebe D. Increase in detection of Corynebacterium diphtheriae in Canada: 2006-2019. Can Commun Dis Rep. 2019;45:296–301. DOIPubMedGoogle Scholar
- Chorlton SD, Ritchie G, Lawson T, Romney MG, Lowe CF. Whole-genome sequencing of Corynebacterium diphtheriae isolates recovered from an inner-city population demonstrates the predominance of a single molecular strain. J Clin Microbiol. 2020;58:e01651–19. DOIPubMedGoogle Scholar
- Dewinter LM, Bernard KA, Romney MG. Human clinical isolates of Corynebacterium diphtheriae and Corynebacterium ulcerans collected in Canada from 1999 to 2003 but not fitting reporting criteria for cases of diphtheria. J Clin Microbiol. 2005;43:3447–9. DOIPubMedGoogle Scholar
- Clinical and Laboratory Standards Institute. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; third edition (M45). Wayne (PA): The Institute; 2015.
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37:540–6. DOIPubMedGoogle Scholar
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–5. DOIPubMedGoogle Scholar
- Hennart M, Crestani C, Bridel S, Armatys N, Brémont S, Carmi-Leroy A, et al. A global Corynebacterium diphtheriae genomic framework sheds light on current diphtheria reemergence. Peer Community J. 2023;3:
e76 . DOIGoogle Scholar - Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. DOIPubMedGoogle Scholar
- Tonkin-Hill G, MacAlasdair N, Ruis C, Weimann A, Horesh G, Lees JA, et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 2020;21:180. DOIPubMedGoogle Scholar
- Argimón S, Abudahab K, Goater RJE, Fedosejev A, Bhai J, Glasner C, et al. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom. 2016;2:
e000093 . DOIPubMedGoogle Scholar - Zhou Z, Alikhan N-F, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, et al. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018;28:1395–404. DOIPubMedGoogle Scholar
1These first authors contributed equally to this article.
2These senior authors contributed equally to this article.