Volume 31, Number 2—February 2025
Dispatch
Diphtheria Toxin–Producing Corynebacterium ramonii in Inner-City Population, Vancouver, British Columbia, Canada, 2019–2023
Abstract
We conducted patient chart reviews and whole-genome sequencing of wound specimens containing presumptive Corynebacterium ulcerans from Vancouver, British Columbia, Canada, during July 2019–July 2023. Sequencing confirmed 8/14 isolates were C. ramonii and identified 2 distinct clusters. Molecular methods should be used to clinically differentiate potential toxin-producing Corynebacterium spp.
Corynebacterium ramonii, a member of the C. diphtheriae species complex, has recently been differentiated from C. ulcerans (1). C. ulcerans is associated with zoonotic transmission (predominantly from infected cats and dogs), whereas C. ramonii is suspected of potential human-to-human transmission; its zoonotic character has not been established (1). C. ulcerans can manifest as respiratory or cutaneous infection similar to C. diphtheriae (2,3). Fourteen isolates have been previously characterized as C. ramonii (1); some harbored the diphtheria toxin gene, underscoring the clinical and public health implications of correctly identifying this organism.
In Canada, where incidence of C. ulcerans infections is low, a 35% increase in toxin testing referrals to the National Microbiology Laboratory in Winnipeg, Manitoba, occurred during 2006–2019; twenty-two cases of C. ulcerans infection were identified (45% were diphtheria toxin positive) (4). Although multiple reports of nontoxigenic cutaneous diphtheria (caused by C. diphtheriae) have occurred in the inner city of Vancouver, British Columbia, only sporadic cases of toxigenic C. ulcerans have been reported (5,6). Because of the additional reported C. ulcerans cases, most of which were subsequently identified as C. ramonii infections, we conducted a clinical, epidemiologic, and genomic review of C. ramonii infections in Vancouver. The University of British Columbia/Providence Health Care Research Institute provided ethics approval for this study (approval no. H22-03695).
St. Paul’s Hospital (SPH) microbiology laboratory and LifeLabs in Vancouver performed a retrospective review of all specimens collected during January 2019–July 2023 that had presumptive C. ulcerans, according to matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry analysis. We extracted clinical data from electronic medical records for patients seen at SPH (no clinical information was available for LifeLabs’ cases), which included microbiology results, hospital course (inpatient/critical care admission), antimicrobial drugs, 30-day mortality, and risk factors for infection (housing status, substance use, and livestock/domestic animal exposure). We characterized bacterial cultures with C. ulcerans by using the MALDI Biotyper sirius System (Bruker, https://www.bruker.com) according to the manufacturer’s recommendations without specific extraction. We analyzed the MALDI-TOF mass spectrometry data by using the Flex Analysis function on the MALDI Biotyper (BDAL version 12, MBT Compass Library version K). At SPH, we performed antimicrobial drug susceptibility testing by using ETEST penicillin, erythromycin, clindamycin, and vancomycin gradient strips (bioMérieux, https://www.biomerieux.com) (7). The National Microbiology Laboratory performed all toxin testing by using PCR and modified Elek tests. For whole-genome sequencing (WGS), we extracted DNA from isolated bacterial colonies obtained from our archives by using the MagNA Pure 24 System (Roche Diagnostics, https://diagnostics.roche.com) and sequenced those on a GridION instrument (Oxford Nanopore Technologies, https://www.nanoporetech.com) by using R10.4.1 flow cells. We performed data acquisition and base calling into fast5 files by using MinKNOW version 23.07.12 and Guppy version 6.4.6 (Oxford Nanopore Technologies) and assembled raw FASTQ files by using Flye version 2.9 (8). We checked genome assembly quality by using QUAST v5.2.0 and analyzed assemblies by using diphtOscan (9,10). We inferred maximum-likelihood phylogeny by using IQ-TREE version 2.3.4 and a general time reversible plus gamma model; we obtained a core gene alignment by using Panaroo version 1.5.0, visualized with Microreact (https://microreact.org/project/rgLWfs1derHFm4K5ShbxgJ-c-ramonii-and-c-ulcerans-vancouver-canada) (11–13). We obtained core genome multilocus sequence typing (cgMLST) profiles by tagging the genomes for known alleles within the BIGSdb-Pasteur database (https://bigsdb.pasteur.fr/diphtheria); we used the cgMLST_ulcerans scheme. We constructed minimum-spanning trees by using GrapeTree directly from the BIGSdb-Pasteur plug-in (14; C. Crestani et al., unpub. data, https://doi.org/10.1101/2024.08.22.609154).
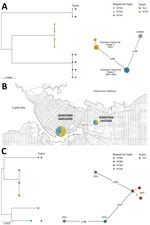
C. ulcerans was initially identified in cultures from 14 patients (SPH, n = 9; LifeLabs, n = 5) by using MALDI-TOF mass spectrometry (scores >2.0). Eight of those samples had a spectral peak at 5405.40 (range 5404–5407) m/z, which is associated with C. ramonii (1). All 14 isolates underwent WGS and genotyping, confirming those 8 isolates were C. ramonii sequence type (ST) 335 (n = 4) and ST341 (n = 4) (Figure, panel A), originating from the inner-city Vancouver (Figure, panel B). The cgMLST results suggested 2 C. ramonii clusters existed; isolates in cluster X had 1 allelic mismatch, and cluster Y had no allelic mismatches (Figure, panel A). We confirmed the remaining 6/14 isolates were C. ulcerans (ST325, ST339, ST669 [3 isolates], and ST895) (Figure, panel C). Those 6 isolates did not originate from a specific neighborhood. All 14 cultures were polymicrobial and recovered from lower extremity wounds (Table).
Three of 8 patients with C. ramonii infection required admission to the hospital (average duration 6 days); critical care admissions were not required, and we observed no 30-day mortality. Toxigenic systemic signs were not observed in the 3 patients infected with diphtheria toxin-producing C. ramonii. Of the 6 patients who had electronic medical records, all were treated with antimicrobial drugs (piperacillin/tazobactam, amoxicillin/clavulanate, trimethoprim/sulfamethoxazole, cephalexin, cefazolin, or ceftriaxone). All 8 patients with C. ramonii infections were associated geographically within downtown Vancouver (Figure, panel B), including 5 persons experiencing homelessness. However, C. ulcerans cases were distributed throughout the city outside of downtown (according to postal codes). All patients with C. ramonii infections reported a history of substance use disorder, and none had documented livestock or domestic animal exposure.
C. ramonii cases clustered exclusively within downtown inner-city Vancouver, whereas C. ulcerans cases occurred outside of the city’s downtown core. Human-to-human transmission of C. ramonii has been hypothesized (1), and our findings provide evidence for possible human-to-human transmission. WGS showed 2 distinct C. ramonii clusters (ST335 and ST341) within the same community. Vancouver’s downtown core has high rates of poverty, persons experiencing homelessness, and substance abuse, and persons might transmit bacteria via close contact, such as that observed for a previous cutaneous C. diphtheriae cluster (5). C. ulcerans infections are associated with zoonotic transmission; however, animal exposures were not observed for patients with C. ramonii infections. C. ramonii was identified in wounds along with other bacteria associated with human reservoirs, such as Streptococcus pyogenes, S. dysgalactiae subspecies equisimilis, and Arcanobacterium hemolyticum. The clinical manifestations of C. ramonii infections align more closely to those of cutaneous C. diphtheriae than to those of C. ulcerans infections (5).
Our study highlights the role for MALDI-TOF mass spectrometry identification of C. ramonii; the initial description of C. ramonii reported a unique peak at 5405.40 m/z (1). In this study, C. ramonii identification required WGS; however, a retrospective review of spectra confirmed the presence of the 5405.40 m/z peak, although the range was broader. MALDI-TOF mass spectrometry is used widely in clinical laboratories, and C. ramonii prevalence can be more accurately estimated when peak analysis of mass spectrograms is performed to avoid misidentification as C. ulcerans. Continual updating of MALDI-TOF mass spectrometry databases is also needed to enable accurate detection of emerging pathogens, such as C. ramonii. Although 16S rRNA gene sequence analysis has been increasingly used in clinical laboratories, it has not been as effective as rpoB gene sequence determinations for differentiating between Corynebacterium spp. (1).
The first limitation of our study is the relatively small number of cases available for analysis. Because wound cultures were polymicrobial, it is unclear to what degree C. ramonii contributed to wound infections. Second, we could not perform a formal case–control study; patients with C. ulcerans infection were primarily seen in outpatient clinics outside of our healthcare network, and complete medical records (including animal contact histories) were not accessible.
In conclusion, our findings correlate with the initial clinical description of C. ramonii, which appears to manifest symptoms similar to those of cutaneous C. diphtheriae infections. Using molecular methods, such as WGS or manual MALDI-TOF mass spectral analysis, will be needed to clinically differentiate between C. ulcerans, C. ramonii, and other members of the C. diphtheriae species complex.
Dr. Lowe is a medical microbiologist and infection prevention and control physician at St. Paul’s Hospital in Vancouver and a clinical professor at the University of British Columbia, British Columbia, Canada. His research interests focus on molecular diagnostics, clinical virology, and the prevention of nosocomial infections.
Acknowledgments
We thank Jennifer Bilawka and Leah Gowland for their technical support and the British Columbia Centre for Disease Control Public Health Laboratory and National Microbiology Laboratory for coordinating and performing toxin tests.
Sequence data for this project are available at https://bigsdb.pasteur.fr/diphtheria, project number 30. Phylogenetic analyses of C. ulcerans and C. ramonii, together with all the relevant metadata, are available in Microreact (https://microreact.org/project/rgLWfs1derHFm4K5ShbxgJ-c-ramonii-and-c-ulcerans-vancouver-canada).
This research was funded, in part, by Institut Pasteur.
References
- Crestani C, Arcari G, Landier A, Passet V, Garnier D, Brémont S, et al. Corynebacterium ramonii sp. nov., a novel toxigenic member of the Corynebacterium diphtheriae species complex. Res Microbiol. 2023;174:
104113 . DOIPubMedGoogle Scholar - Otshudiema JO, Acosta AM, Cassiday PK, Hadler SC, Hariri S, Tiwari TSP. Respiratory illness caused by Corynebacterium diphtheriae and C. ulcerans, and use of diphtheria antitoxin in the United States, 1996–2018. Clin Infect Dis. 2021;73:e2799–806. DOIPubMedGoogle Scholar
- Moore LSP, Leslie A, Meltzer M, Sandison A, Efstratiou A, Sriskandan S. Corynebacterium ulcerans cutaneous diphtheria. Lancet Infect Dis. 2015;15:1100–7. DOIPubMedGoogle Scholar
- Bernard KA, Pacheco AL, Burdz T, Wiebe D. Increase in detection of Corynebacterium diphtheriae in Canada: 2006-2019. Can Commun Dis Rep. 2019;45:296–301. DOIPubMedGoogle Scholar
- Chorlton SD, Ritchie G, Lawson T, Romney MG, Lowe CF. Whole-genome sequencing of Corynebacterium diphtheriae isolates recovered from an inner-city population demonstrates the predominance of a single molecular strain. J Clin Microbiol. 2020;58:e01651–19. DOIPubMedGoogle Scholar
- Dewinter LM, Bernard KA, Romney MG. Human clinical isolates of Corynebacterium diphtheriae and Corynebacterium ulcerans collected in Canada from 1999 to 2003 but not fitting reporting criteria for cases of diphtheria. J Clin Microbiol. 2005;43:3447–9. DOIPubMedGoogle Scholar
- Clinical and Laboratory Standards Institute. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; third edition (M45). Wayne (PA): The Institute; 2015.
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37:540–6. DOIPubMedGoogle Scholar
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–5. DOIPubMedGoogle Scholar
- Hennart M, Crestani C, Bridel S, Armatys N, Brémont S, Carmi-Leroy A, et al. A global Corynebacterium diphtheriae genomic framework sheds light on current diphtheria reemergence. Peer Community J. 2023;3:
e76 . DOIGoogle Scholar - Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. DOIPubMedGoogle Scholar
- Tonkin-Hill G, MacAlasdair N, Ruis C, Weimann A, Horesh G, Lees JA, et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 2020;21:180. DOIPubMedGoogle Scholar
- Argimón S, Abudahab K, Goater RJE, Fedosejev A, Bhai J, Glasner C, et al. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom. 2016;2:
e000093 . DOIPubMedGoogle Scholar - Zhou Z, Alikhan N-F, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, et al. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018;28:1395–404. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleOriginal Publication Date: January 07, 2025
1These first authors contributed equally to this article.
2These senior authors contributed equally to this article.
Table of Contents – Volume 31, Number 2—February 2025
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Christopher F. Lowe, St. Paul’s Hospital, 1081 Burrard St, Vancouver, BC V6Z 1Y6, Canada
Top