Volume 31, Number 3—March 2025
Dispatch
Outbreak Caused by Multidrug-Resistant Mycobacterium Tuberculosis with Unusual Combination of Resistance Mutations, Northern Argentina, 2006–2022
Figure 1
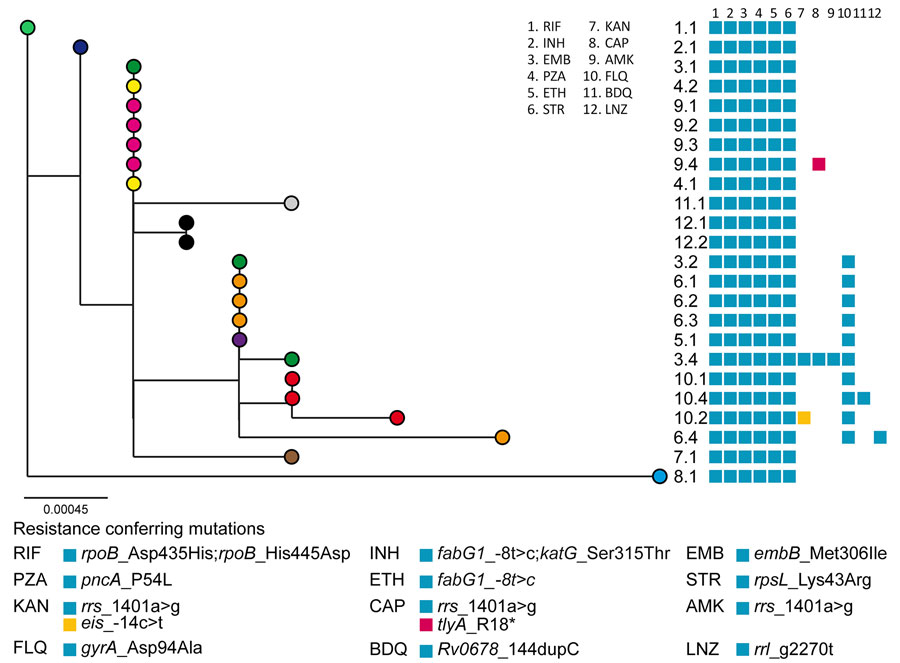
Figure 1. Maximum-likelihood phylogenetic tree of the 24 isolates from the MDR Mycobacterium tuberculosis outbreak strain Ch, Chaco, Argentina, 2006–2022, analyzed by whole-genome sequencing. Each patient is represented by a colored dot. Labels indicate the patient and isolate identification numbers. Blocks indicate the resistance-conferring mutations detected in each isolate, as indicated below the tree. Scale bar indicates number of substitutions per variable site. MDR, multidrug-resistant.
1These first authors contributed equally to this article.
Page created: February 04, 2025
Page updated: March 05, 2025
Page reviewed: March 05, 2025
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.