Volume 31, Number 4—April 2025
Research
Attribution of Salmonella enterica to Food Sources by Using Whole-Genome Sequencing Data
Figure 3
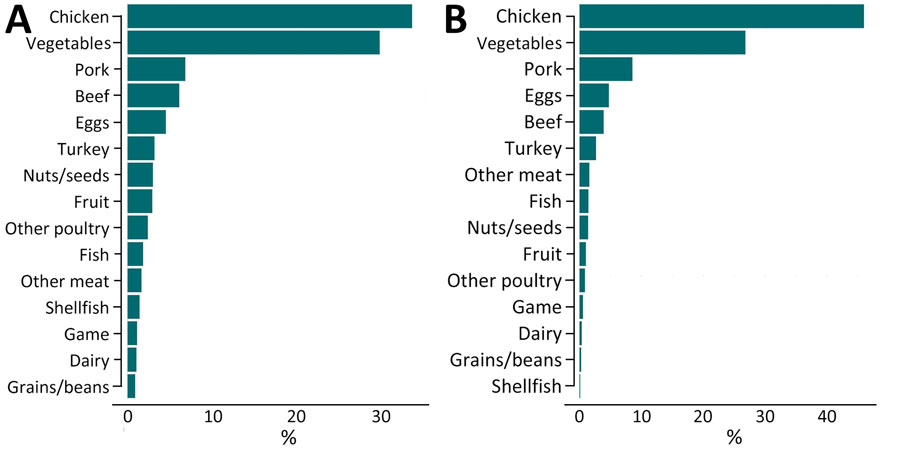
Figure 3. Predictions from a random forest model of sources of US human illnesses among patients without reported history of international travel who had whole-genome sequenced Salmonella isolates reported to the Foodborne Diseases Active Surveillance Network, 2014–2017. A) Predictions of sources of human illnesses among all patients (N = 6,470). B) Predictions of food category sources of human illnesses among patients renormalized among isolates with a >0.50 probability of attribution to a single food category (n = 3,686).
1These first authors contributed equally to this article.
Page created: March 02, 2025
Page updated: March 24, 2025
Page reviewed: March 24, 2025
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.