Volume 31, Number 5—May 2025
Dispatch
Molecular Detection of Histoplasma in Bat-Inhabited Tunnels of Camino de Hierro Tourist Route, Spain
Figure
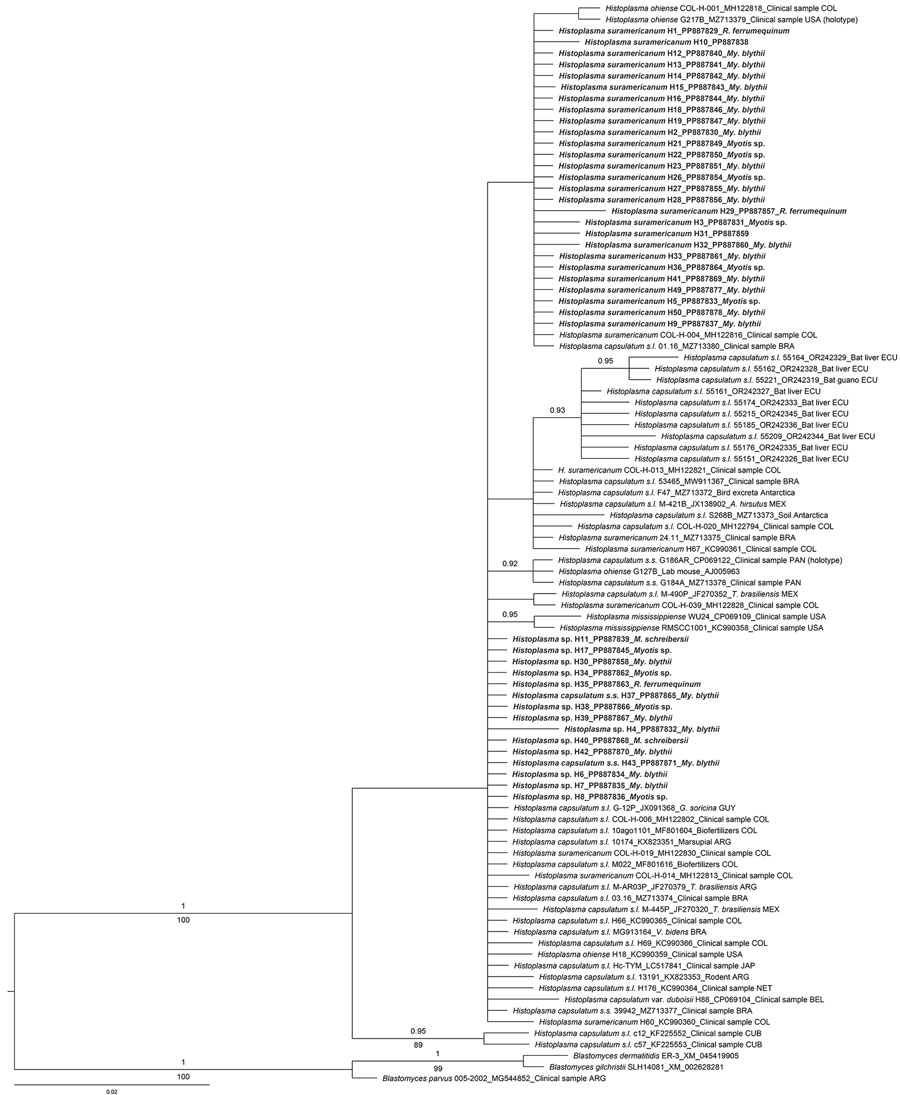
Figure. Bayesian tree based on 210-bp partial Hcp100 gene sequences obtained from 42 guano samples collected in the tunnels of the Camino de Hierro tourist route in study of molecular detection of Histoplasma, Spain. Representatives of the genus Blastomyces were used as outgroup. GenBank reference isolates are labeled by species name, specimen code, accession number, host or source (if known), and 3-letter country code. Bold text indicates sequences obtained in this study, labeled by taxon name, isolate code, accession number, and source bat species. Numbers above the branches correspond to Bayesian posterior probability and numbers below the branches to maximum-likelihood bootstrap values; values are shown if posterior probability is >0.90 and bootstrap value is >75%. Scale bar indicates average number of substitutions per site.
1These first authors contributed equally to this article.