Volume 31, Number 5—May 2025
Dispatch
High Prevalence of Influenza D Virus Infection in Swine, Northern Ireland
Figure 2
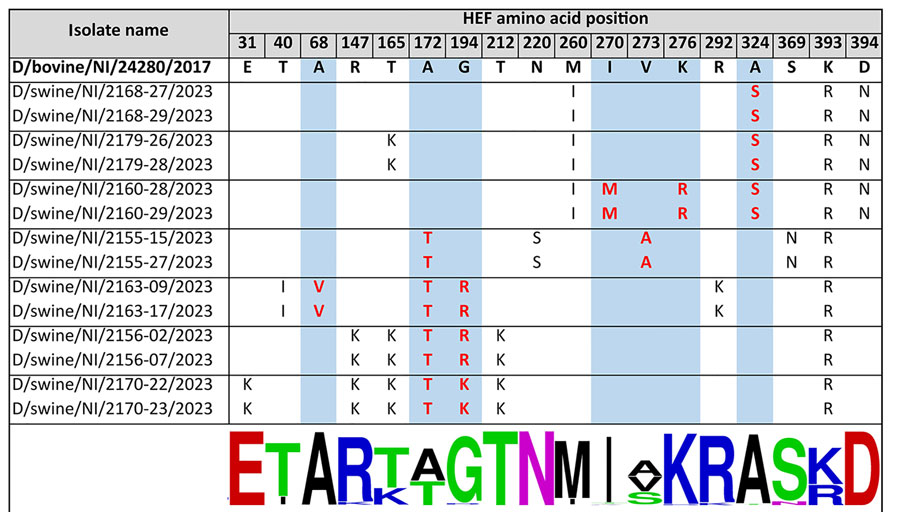
Figure 2. Amino acid substitutions in swine-origin influenza D virus HEF protein relative to bovine-origin influenza D virus, Northern Ireland. Changes relative to the bovine-origin sequence are displayed. Amino acid numbering is according to Song et al. (6). Substitutions occurring at the receptor-binding site and esterase are highlighted with red text on a blue background. Sequence conservation at each position is indicated by logo generated at WebLogo (https://weblogo.berkeley.edu/logo.cgi) (bottom) on the basis of alignment of 169 influenza D virus sequences. HEF, hemagglutinin-esterase fusion; NI, Northern Ireland.
References
- Kwasnik M, Rola J, Rozek W. Influenza D in domestic and wild animals. Viruses. 2023;15:2433. DOIPubMedGoogle Scholar
- Gaudino M, Chiapponi C, Moreno A, Zohari S, O’Donovan T, Quinless E, et al. Evolutionary and temporal dynamics of emerging influenza D virus in Europe (2009-22). Virus Evol. 2022;8:
veac081 . DOIPubMedGoogle Scholar - Dane H, Duffy C, Guelbenzu M, Hause B, Fee S, Forster F, et al. Detection of influenza D virus in bovine respiratory disease samples, UK. Transbound Emerg Dis. 2019;66:2184–7. DOIPubMedGoogle Scholar
- Hause BM, Ducatez M, Collin EA, Ran Z, Liu R, Sheng Z, et al. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 2013;9:
e1003176 . DOIPubMedGoogle Scholar - Lagan P, Hamil M, Cull S, Hanrahan A, Wregor RM, Lemon K. Swine influenza A virus infection dynamics and evolution in intensive pig production systems. Virus Evol. 2024;10:
veae017 . DOIPubMedGoogle Scholar - Song H, Qi J, Khedri Z, Diaz S, Yu H, Chen X, et al. An open receptor-binding cavity of hemagglutinin-esterase-fusion glycoprotein from newly identified influenza D virus: basis for its broad cell tropism. PLoS Pathog. 2016;12:
e1005411 . DOIPubMedGoogle Scholar - Gorin S, Richard G, Hervé S, Eveno E, Blanchard Y, Jardin A, et al. Characterization of influenza D virus reassortant strain in swine from mixed pig and beef farm, France. Emerg Infect Dis. 2024;30:1672–6. DOIPubMedGoogle Scholar
- Kaplan BS, Falkenberg S, Dassanayake R, Neill J, Velayudhan B, Li F, et al. Virus strain influenced the interspecies transmission of influenza D virus between calves and pigs. Transbound Emerg Dis. 2021;68:3396–404. DOIPubMedGoogle Scholar
- Gorin S, Richard G, Quéguiner S, Chastagner A, Barbier N, Deblanc C, et al. Pathogenesis, transmission, and within-host evolution of bovine-origin influenza D virus in pigs. Transbound Emerg Dis. 2024;2024:
9009051 . DOIGoogle Scholar
Page created: March 11, 2025
Page updated: April 25, 2025
Page reviewed: April 25, 2025
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.