Genomic Characterization of Leishmania tropica in Cutaneous Leishmaniasis, Somali Region, Ethiopia, 2023
Adugna Abera
1
, Pieter Monsieurs
1, Myrthe Pareyn, Dereje Beyene, Geremew Tasew, Allison Aroni-Soto, Mahlet Belachew, Desalegn Geleta, Bethlehem Adnew, Bokretsion Gidey, Henok Tadesse, Atsbeha Gebreegziabxier, Rajiha Abubeker, Abraham Ali, Ketema Tafess, Tobias F. Rinke de Wit, Johan van Griensven, Jean-Claude Dujardin, Dawit Wolday
2, and Malgorzata Anna Domagalska
2
Author affiliation: College of Natural and Computational Sciences, Addis Ababa University, Addis Ababa, Ethiopia (A. Abera, D. Beyene); Ethiopian Public Health Institute, Addis Ababa (A. Abera, G. Tasew, M. Belachew, D. Geleta, B. Gidey, H. Tadesse, A. Gebreegziabxier, R. Abubeker, A. Ali, D. Wolday); Institute of Tropical Medicine, Antwerp, Belgium (P. Monsieurs, M. Pareyn, A. Aroni-Soto, J. van Griensven, J.-C. Dujardin, M.A. Domagalska); Armauer Hansen Research Institute, Addis Ababa (B. Adnew); Adama Science and Technology University, Adama, Ethiopia (K. Tafess); Amsterdam Institute for Global Health and Development, Amsterdam University Medical Centre, University of Amsterdam, Amsterdam, the Netherlands (T.F.R. de Wit); Michael G. DeGroote Institute for Infectious Disease Research, Hamilton, Ontario, Canada (D. Wolday); McMaster Immunology Research Center, Health Sciences, McMaster University, Hamilton (D. Wolday)
Main Article
Figure 2
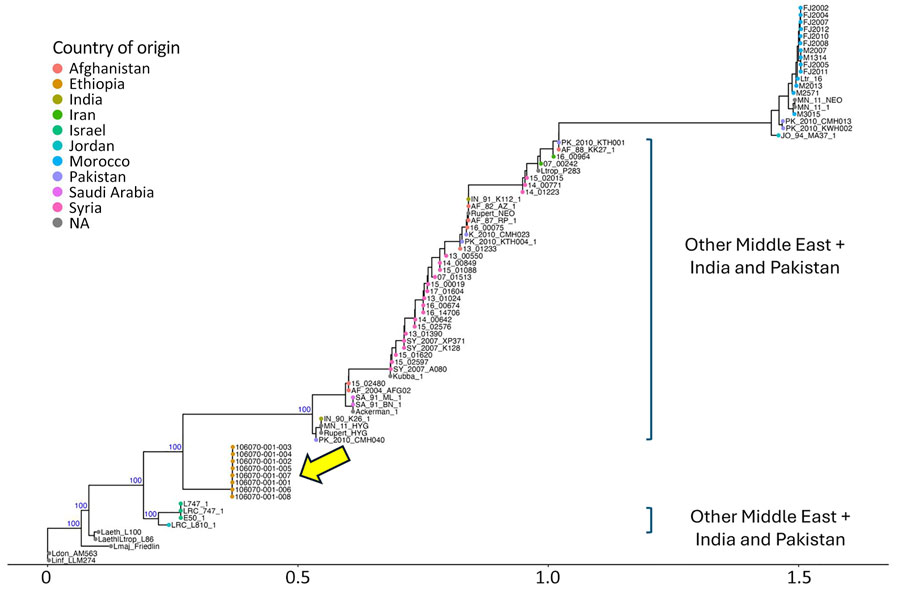
Figure 2. Rooted phylogenetic tree for genomic characterization of Leishmania tropica in cutaneous leishmaniasis, Somali region, Ethiopia, 2023. Phylogenetic tree of all publicly available L. tropica genomes (Appendix Table 3) was generated with RAxML (https://github.com/amkozlov/raxml-ng) with the general time reversible and gamma substitution model and showing close clustering of the 8 samples from Ethiopia (yellow arrow). Blue text indicates crucial bootstrap values. The L. infantum LLM274 genome was included as an outgroup. Scale bar indicates number of single-nucleotide polymorphism differences. NA, geographic origin of the L. tropica sample is unknown.
Main Article
Page created: May 31, 2025
Page updated: June 25, 2025
Page reviewed: June 25, 2025
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
