Volume 31, Number 7—July 2025
Research Letter
Genomic Characterization of Leishmania tropica in Cutaneous Leishmaniasis, Somali Region, Ethiopia, 2023
Cite This Article
Citation for Media
Abstract
We sequenced Leishmania tropica genomes from 8 human skin samples collected in a newly emerging focus of cutaneous leishmaniasis in the Somali region of Ethiopia. We found a variant with unique genomic signatures of drug resistance. Public health officials should use genomic surveillance to slow expansion of L. tropica.
Since the successful Kala-Azar elimination program in the Indian subcontinent, the hotspot of worldwide leishmaniasis has moved to East Africa (1). Among the different affected countries, Ethiopia deserves particular attention, given the heterogeneous eco-epidemiology of leishmaniasis, its clinical polymorphism, and the complex taxonomy of Leishmania tropica parasites. The disease is endemic in different biotopes from lowlands to highlands, and transmission involves different hosts and vectors (2). The 4 major clinical forms of leishmaniasis are visceral leishmaniasis (VL), causing 2,500–4,000 reported cases, and 3 forms of cutaneous leishmaniasis (CL), localized, diffuse, and muco-cutaneous, causing ≈50,000 reported cases. L. donovani (VL and occasionally CL) and L. aethiopica (all 3 CL forms) are the most reported species, and L. tropica (CL) was isolated once from a human patient (2); several interspecies hybrids have been observed (3).
The epidemiology of the disease is affected by human migration and displacement because of famine and regular conflicts in the country and by environmental changes. We previously highlighted the need for genomic surveillance of leishmaniasis by using highly sensitive, resolutive, and untargeted whole-genome sequencing (WGS) methods (4) for the following reasons: since the discovery of hybridization and genetic introgression (5), robust species identification should theoretically be on the basis of multigenic approaches covering several regions of the genome (and not only single gene approaches); WGS is needed to assess the genetic similarity among parasites sampled from different patients, thereby confirming the outbreak nature of a focus; and WGS can be used to find signatures of drug resistance and guide patient management.
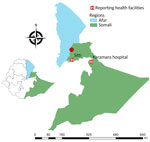
In August 2023, an outbreak of CL was detected among immunologically naive militia recently deployed in the eastern Somali region of Ethiopia. That area did not have a previous history of CL, but sporadic cases of VL were reported. The clinical manifestations of the CL cases (multiple wet lesions) was not comparable to what is typically observed in Ethiopia and neighboring countries (single dry lesions). Hsp70 amplicon sequencing identified the CL pathogen as L. tropica (A. Abera et al., unpub. data, https://www.medrxiv.org/content/10.1101/2024.10.05.24314933v1).
We undertook a more in-depth molecular characterization of L. tropica samples collected in the focus in the Somali region (Figure 1). We used direct genome sequencing of Leishmania in host tissues (SureSelect sequencing; Agilent Technologies, https://www.agilent.com) that did not require parasite isolation and cultivation (6). That method was previously validated for L. donovani in bone marrow (6) and blood samples (7), and we used it for the first time for skin samples from CL patients.
We submitted 8 of the CL samples from the Somali region for SureSelect sequencing by using a capture panel of probes designed for the L. aethiopica genome. SureSelect sequencing should work well with phylogenetically related species such as L. tropica. We used competitive mapping (Appendix Figure 1) and phylogenetic analysis (Appendix Figure 2) for the species identification of the 8 samples. Those samples (Appendix Figure 2, yellow arrow) clearly branch in the L. tropica cluster and are genetically very different from L. aethiopica, L. major, L. donovani, and interspecies hybrids.
In a second step, we only focused on L. tropica genomes (Figure 2). That focus provided 4 major insights. The 8 Somali region parasites constitute a L. tropica variant not previously reported in analyzed genomes. Those variants form a distinct cluster separate from genotypes reported thus far from Israel and Jordan and other Middle East variants. The L. tropica variants cluster together and are genetically homogeneous (on average, 122 single-nucleotide polymorphisms between samples), consistent with an outbreak-related scenario. We also found homozygous missense mutations or frameshifts in 14 genes reported to be involved in drug resistance, mostly antimony (Appendix Figure 3). That signature has not previously been reported in the L. tropica genome, which confirms the unique character of the samples from this region. Further work is required to understand the clinical effects of the discovery.
L. tropica is essentially endemic in Morocco, Turkey, Syria, Israel, Iraq, Azerbaijan, Iran, Uzbekistan, Afghanistan, Pakistan, and India (8). The broad distribution of L. tropica likely results from the anthroponotic nature of L. tropica transmission and the old communication axes in many of those countries, such as trade routes. In some regions, sporadic cases are reported, and the disease is thought to be zoonotic; possible animal reservoirs included hyraxes, bats, or wild rodents (9). The high genomic homogeneity in our sampled population shows the occurrence of an L. tropica outbreak in the Somali region of Ethiopia. We do not have the ability to trace whether the origin of the outbreak was a primary human case from which the parasite population spread or an animal reservoir. Nevertheless, this study highlights the risk for further expansion of the parasites from the human cases in the focus in the Somali region of Ethiopia. Public health officials should use genomic surveillance in humans, insect vectors, and animals in Ethiopia and neighboring countries, such as Kenya, where L. tropica was recently reported (10), to slow expansion of L. tropica.
Dr. Abera is a senior researcher at the Ethiopian Public Health Institute. His research focuses on molecular epidemiology, drug resistance, diagnostics, and metagenomics research applied to malaria, neglected tropical diseases, and arboviral diseases in Ethiopia.
Acknowledgments
This article was preprinted at https://doi.org/10.1101/2024.10.15.617758.
Genomic sequence reads of the parasites from the 8 samples have been submitted to the National Center for Biotechnology Information Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra; Bioproject no. PRJNA1172382).
This study was financially supported by the Belgian Directorate-General for Development Cooperation Framework Agreement 5 Ethiopia program (awarded to J.v.G. and G.T.), the Dioraphte Foundation (Spatial-CL, project no. CFP-RD2020 20020401), EpiGen-Ethiopia (project no. 101103188, funded through the Global Health EDCTP3 program–European Union), and the Flemish Ministry of Science and Innovation (to M.A.D.).
References
- Alvar J, Beca-Martínez MT, Argaw D, Jain S, Aagaard-Hansen J. Social determinants of visceral leishmaniasis elimination in Eastern Africa. BMJ Glob Health. 2023;8:
e012638 . DOIPubMedGoogle Scholar - Hailu A, Di Muccio T, Abebe T, Hunegnaw M, Kager PA, Gramiccia M. Isolation of Leishmania tropica from an Ethiopian cutaneous leishmaniasis patient. Trans R Soc Trop Med Hyg. 2006;100:53–8. DOIPubMedGoogle Scholar
- Hadermann A, Heeren S, Maes I, Dujardin JC, Domagalska MA, Van den Broeck F. Genome diversity of Leishmania aethiopica. Front Cell Infect Microbiol. 2023;13:
1147998 . DOIPubMedGoogle Scholar - Domagalska MA, Dujardin JC. Next-generation molecular surveillance of TriTryp diseases. Trends Parasitol. 2020;36:356–67. DOIPubMedGoogle Scholar
- Lypaczewski P, Matlashewski G. Leishmania donovani hybridisation and introgression in nature: a comparative genomic investigation. Lancet Microbe. 2021;2:e250–8. DOIPubMedGoogle Scholar
- Domagalska MA, Imamura H, Sanders M, Van den Broeck F, Bhattarai NR, Vanaerschot M, et al. Genomes of Leishmania parasites directly sequenced from patients with visceral leishmaniasis in the Indian subcontinent. PLoS Negl Trop Dis. 2019;13:
e0007900 . DOIPubMedGoogle Scholar - Monsieurs P, Cloots K, Uranw S, Banjara MR, Ghimire P, Burza S, et al. Source tracing of Leishmania donovani in emerging foci of visceral leishmaniasis, Western Nepal. Emerg Infect Dis. 2024;30:611–3. DOIPubMedGoogle Scholar
- World Health Organization. Control of the leishmaniases: WHO TRS N°949 [cited 2024 Sep 11]. https://www.who.int/publications/i/item/WHO-TRS-949
- Kassahun A, Sadlova J, Benda P, Kostalova T, Warburg A, Hailu A, et al. Natural infection of bats with Leishmania in Ethiopia. Acta Trop. 2015;150:166–70. DOIPubMedGoogle Scholar
- Owino BO, Matoke-Muhia D, Alraey Y, Mwangi JM, Ingonga JM, Ngumbi PM, et al. Association of Phlebotomus guggisbergi with Leishmania major and Leishmania tropica in a complex transmission setting for cutaneous leishmaniasis in Gilgil, Nakuru county, Kenya. PLoS Negl Trop Dis. 2019;13:
e0007712 . DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: June 20, 2025
1These authors contributed equally to this article.
2These senior authors contributed equally to this article.
Table of Contents – Volume 31, Number 7—July 2025
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Adugna Abera, Ethiopian Public Health Institute, Arbagnoch St, PO Box 1242, Addis Ababa, Ethiopia
Top