Volume 31, Number 7—July 2025
Dispatch
Detection of Novel Orthobunyavirus Reassortants in Fatal Neurologic Case in Horse and Culicoides Biting Midges, South Africa
Figure 3
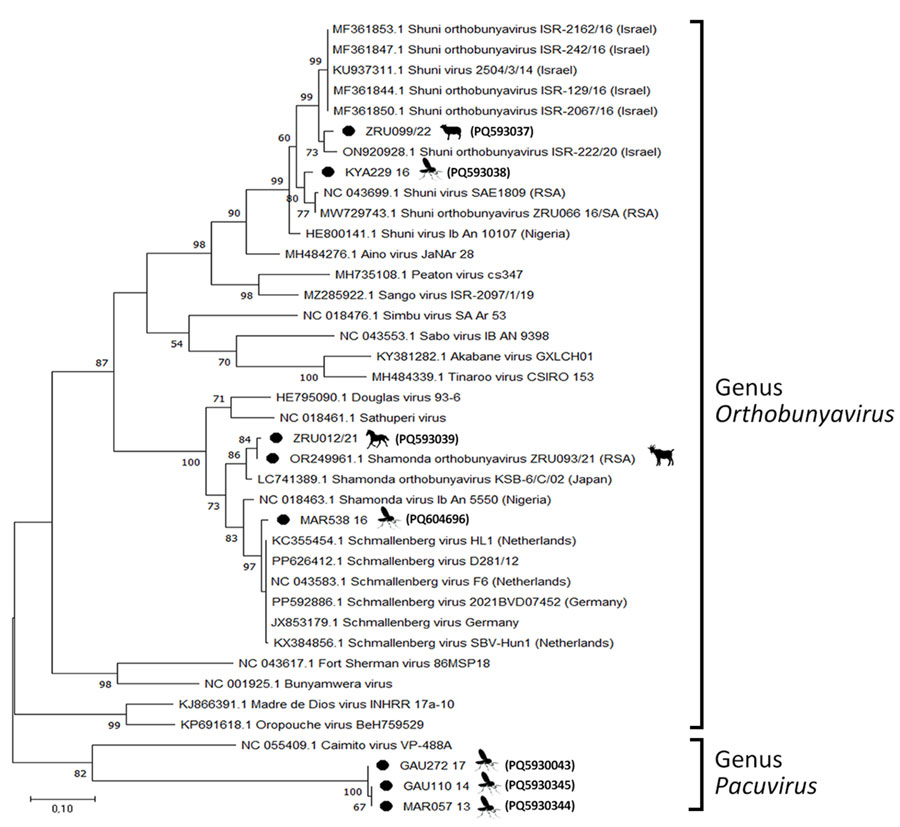
Figure 3. Phylogenetic analysis of Orthobunyavirus L segments in study of reassortants in fatal neurologic case in horse and Culicoides biting midges, South Africa. Tree shows phylogenetic analysis of the positive animal cases and Culicoides pools for the L segment of sequences from South Africa (black dots) relative to orthobunyavirus reference strains available in GenBank within the Simbu serogroup. Tree was constructed in MEGA X (https://www.megasoftware.net) by using the Tamura-Nei model. Bootstrap values from 1,000 replicates are shown at each node. Pacuvirus genus was included as an outgroup to root the tree. Bolded accession numbers in parentheses indicate sequences from this study that were submitted to GenBank. Scale bar indicates nucleotide substitutions per site.