Plasmodium knowlesi Malaria in Persons Returning to Israel from Thailand, 2023
Yael Paran, Ami Neuberger, Muna Massarwa, Maisam Amar, Julia Vainer, Moran Szwarcwort Cohen, Ohad Shalom, Shirly Elbaz, Maya Davidovich, Oscar David Kirstein, and Tamar Grossman

Author affiliation: Tel Aviv Sourasky Medical Center, Tel Aviv, Israel (Y. Paran); Division of Internal Medicine, Rambam Health Care Campus, Haifa, Israel (A. Neuberger, M. Massarwa); Technion Faculty of Medicine, Haifa (A. Neuberger, M. Amar); Infectious Disease Unit, Lady Davis Carmel Medical Center, Haifa (M. Amar); Public Health Laboratories–Jerusalem, Public Health Services, Ministry of Health, Jerusalem, Israel (J. Vainer, S. Elbaz, M. Davidovich, O.D. Kirstein, T. Grossman); Microbiology Laboratory, Rambam Health Care Campus, Haifa (M.S. Cohen); Microbiology Laboratory, Tel Aviv Sourasky Medical Center, Tel Aviv (O. Shalom)
Main Article
Figure 2
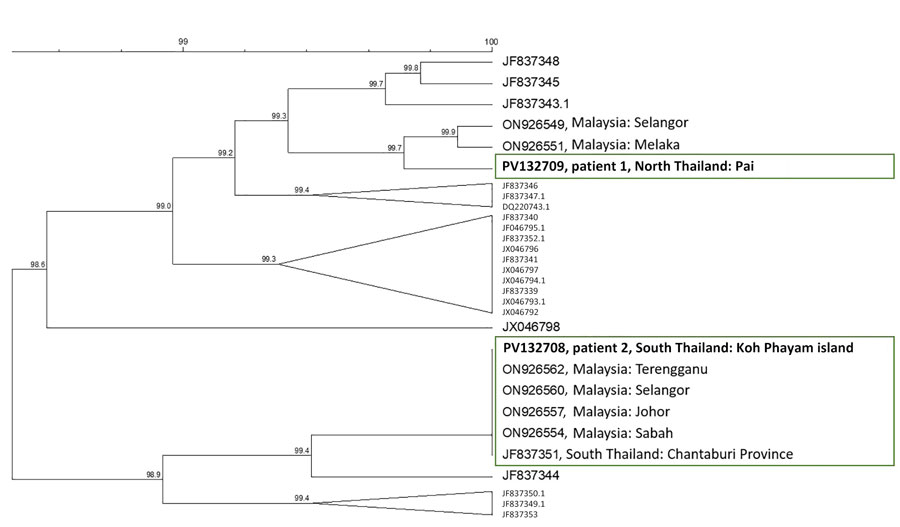
Figure 2. Phylogenetic analysis of Pkmsp1 sequences for Plasmodium knowlesi malaria in persons returning to Israel from Thailand, 2023. Dendogram was based on an 850-bp fragment of the C-terminal region of Pkmsp1, including all published sequences from Thailand, the isolates from patients 1 and 2 in this study (bold text), and the most closely related sequences identified through BLAST (https://blast.ncbi.nlm.nih.gov) analysis. No exact matches were identified for patient 1 (northern Thailand); the geographic origin of 2 closely related sequences is indicated. For patient 2 (southern Thailand), several identical sequences were found in GenBank (green box). Accession numbers are given. Clustering analysis was performed by using the unweighted pair group with arithmetic mean method with open gap penalty set to 100% and unit gap penalty set to 0%, without correction. Scale bar indicates percentage similarity between sequences.
Main Article
Page created: May 16, 2025
Page updated: June 25, 2025
Page reviewed: June 25, 2025
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
