Multidisciplinary Tracking of Highly Pathogenic Avian Influenza A(H5N1) Outbreak in Griffon Vultures, Southern Europe, 2022
Julien Hirschinger, Ursula Höfle, Alberto Sánchez-Cano, Claire Guinat, Guillaume Croville, Marta Barral, José Antonio Donázar, Chloé Le Gall Ladevèze, Mathilda Walch, Vega Alvarez, Xeider Gerrikagoitia, Louis Du Plessis, Simon Dellicour, Eneko Arrondo, José Antonio Sánchez-Zapata, Ainara Cortés-Avizanda, Sara Minayo Martín, Jérémy Tornos, Samuel Perret, Thierry Boulinier, Pascal Orabi, Anne Van De Wiele, Jean-Luc Guerin, Olivier Duriez, and Guillaume Le Loc’h

Author affiliation: IHAP, ENVT, INRAE, Université de Toulouse, Toulouse, France (J. Hirschinger, C. Guinat, G. Croville, C. Le Gall Ladevèze, M. Walch, J.-L. Guerin, G. Le Loc’h); Grupo SaBio, Instituto de Investigación en Recursos Cinegéticos (CSIC-UCLM-JCCM), Ciudad Real, Spain (U. Höfle, A. Sánchez-Cano, S. Minayo Martín); NEIKER–Basque Institute for Agricultural Research and Development, Basque Research and Technology Alliance (BRTA), Derio, Spain (M. Barral, V. Alvarez, X. Gerrikagoitia); Estación Biológica de Doñana, CSIC, Sevilla, Spain (J.A. Donázar, A. Cortés-Avizanda); ETH Zurich, Basel, Switzerland (L. Du Plessis); Swiss Institute of Bioinformatics, Lausanne, Switzerland (L. Du Plessis); Spatial Epidemiology Lab, Université Libre de Bruxelles, Brussels, Belgium (S. Dellicour); Rega Institute, KU Leuven, Leuven, Belgium (S. Dellicour); Universidad Miguel Hernández, Elche, Spain (J.A. Sánchez-Zapata); Centre d’Ecologie Fonctionnelle et Evolutive, Univ Montpellier, CNRS, EPHE, IRD, Montpellier, France (J. Tornos, S. Perret, T. Boulinier, O. Duriez); Ligue pour la Protection des Oiseaux, Rochefort, France (P. Orabi); Office Français de la Biodiversité, Vincennes, France (A. Van De Wiele); University of Granada, Granada, Spain (E. Arrondo)
Main Article
Figure 2
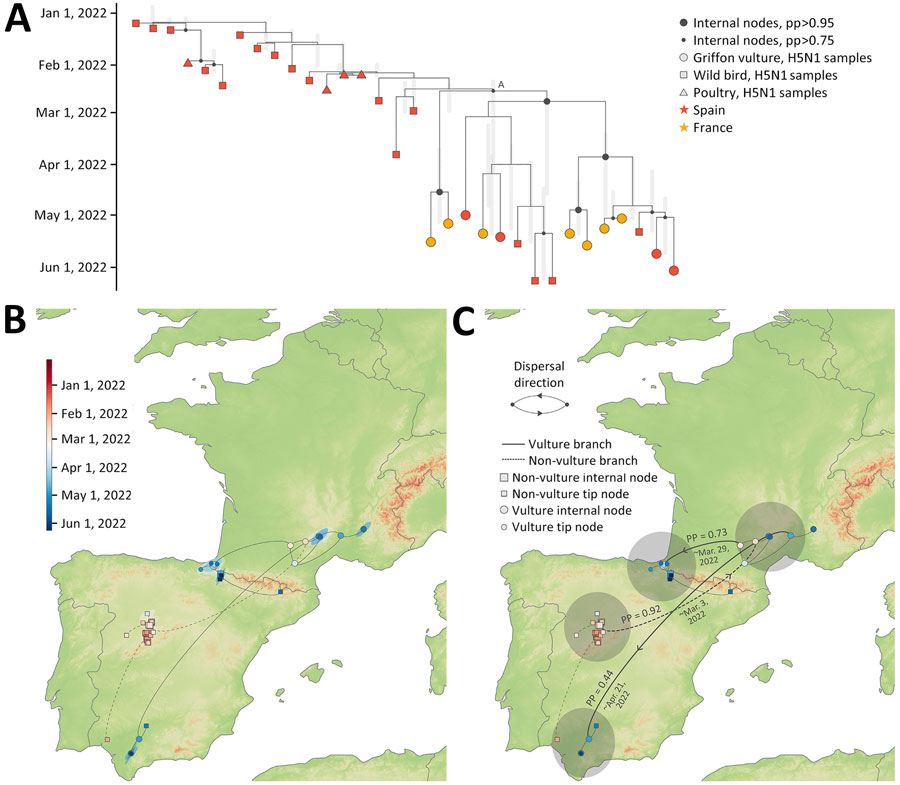
Figure 2. Phylogenetic and phylogeographic analysis conducted in study of multidisciplinary tracking of highly pathogenic avian influenza A(H5N1) outbreak in griffon vultures (Gyps fulvus), southern Europe, 2022. A) Maximum clade credibility (MCC) tree obtained from the time-scaled phylogenetic analysis based on genetic sequences of the hemagglutinin gene segment collected from H5N1 virus–infected birds during November 8, 2021–September 1, 2022, in Spain and France. Vertical light gray bars reflect 95% highest posterior density (HPD) intervals associated with the inferred age of internal nodes. B) Continuous phylogeographic reconstruction of the dispersal history of viral lineages. Specifically, we first report the mapped MCC tree and 80% HPD regions reflecting the uncertainty related to the Bayesian continuous phylogeographic inference; both the MCC tree and HPD regions are based on 1,000 trees sampled from the posterior distribution and colored according to their time of occurrence. Phylogenetic branches associated with the vulture subclade (node A) are displayed as solid lines, whereas dashed lines represent other branches in the clade. C) MCC tree as reported in panels A and B, but this time along the PPs and mean estimates associated with lineage dispersal events that occurred between the 4 main regions involved in the continuous phylogeographic reconstruction. PP, posterior probability.
Main Article
Page created: June 16, 2025
Page updated: July 21, 2025
Page reviewed: July 21, 2025
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
