Volume 31, Number 8—August 2025
Research Letter
Complete Genome Analysis of African Swine Fever Virus Isolated from Wild Boar, India, 2021
Figure 1
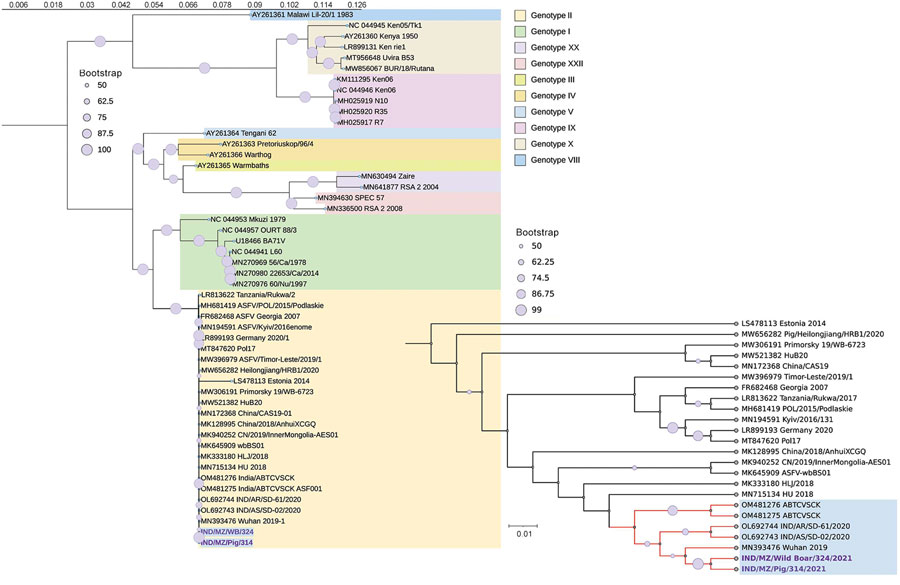
Figure 1. Phylogenetic tree generated from complete genome analysis of ASFV isolated from wild boar and domestic pig, India, 2021. The maximum-likelihood tree was MAFFT aligned (https://mafft.cbrc.jp/alignment/software) by using a general time-reversible plus gamma model in RAxMLGUI version 2.0 (https://sourceforge.net/projects/raxmlgui) and shows the relationship between ASFVs from India and other ASFVs including p72 genotype II. Enlarged area at bottom right shows detail of isolates from this study (bold text) and close reference sequences. GenBank accession numbers are provided. Scale bar indicates the number of expected substitutions per site (tree rooted at mid-point). ASFV, African swine fever virus.
1These authors contributed equally to this article.