Volume 7, Number 2—April 2001
THEME ISSUE
4th Decennial International Conference on Nosocomial and Healthcare-Associated Infections
State of the Art
Molecular Approaches to Diagnosing and Managing Infectious Diseases: Practicality and Costs
Cite This Article
Citation for Media
Abstract
As molecular techniques for identifying and detecting microorganisms in the clinical microbiology laboratory have become routine, questions about the cost of these techniques and their contribution to patient care need to be addressed. Molecular diagnosis is most appropriate for infectious agents that are difficult to detect, identify, or test for susceptibility in a timely fashion with conventional methods.
The tools of molecular biology have proven readily adaptable for use in the clinical diagnostic laboratory and promise to be extremely useful in diagnosis, therapy, and epidemiologic investigations and infection control (1,2). Although technical issues such as ease of performance, reproducibility, sensitivity, and specificity of molecular tests are important, cost and potential contribution to patient care are also of concern (3). Molecular methods may be an improvement over conventional microbiologic testing in many ways. Currently, their most practical and useful application is in detecting and identifying infectious agents for which routine growth-based culture and microscopy methods may not be adequate (4–7).
Nucleic acid-based tests used in diagnosing infectious diseases use standard methods for isolating nucleic acids from organisms and clinical material and restriction endonuclease enzymes, gel electrophoresis, and nucleic acid hybridization techniques to analyze DNA or RNA (6). Because the target DNA or RNA may be present in very small amounts in clinical specimens, various signal amplification and target amplification techniques have been used to detect infectious agents in clinical diagnostic laboratories (5,6). Although mainly a research tool, nucleic acid sequence analysis coupled with target amplification is clinically useful and helps detect and identify previously uncultivatable organisms and characterize antimicrobial resistance gene mutations, thus aiding both diagnosis and treatment of infectious diseases (5,8,9). Automation and high-density oligonucleotide probe arrays (DNA chips) also hold great promise for characterizing microbial pathogens (6).
Although most clinicians and microbiologists enthusiastically welcome the new molecular tests for diagnosing infectious disease, the high cost of these tests is of concern (3). Despite the probability that improved patient outcome and reduced cost of antimicrobial agents and length of hospital stay will outweigh the increased laboratory costs incurred through the use of molecular testing, such savings are difficult to document (3,10,11). Much of the justification for expenditures on molecular testing is speculative (11); however, the cost of equipment, reagents, and trained personnel is real and substantial, and reimbursement issues are problematic (3,11). Given these concerns, a facility's need for molecular diagnostic testing for infectious diseases should be examined critically by the affected clinical and laboratory services. In many instances, careful overseeing of test ordering and prudent use of a reference laboratory may be the most viable options.
Commercial kits for the molecular detection and identification of infectious pathogens have provided a degree of standardization and ease of use that has facilitated the introduction of molecular diagnostics into the clinical microbiology laboratory (Table 1). The use of nucleic acid probes for identifying cultured organisms and for direct detection of organisms in clinical material was the first exposure that most laboratories had to commercially available molecular tests. Although these probe tests are still widely used, amplification-based methods are increasingly employed for diagnosis, identification and quantitation of pathogens, and characterization of antimicrobial-drug resistance genes. Commercial amplification kits are available for some pathogens (Table 1), but some clinically important pathogens require investigator-designed or "home-brew" methods (Table 2). In addition, molecular strain typing, or genotyping, has proven useful in guiding therapeutic decisions for certain viral pathogens and for epidemiologic investigation and infection control (2,12).
Commercial kits containing non-isotopically labeled nucleic acid probes are available for direct detection of pathogens in clinical material and identification of organisms after isolation in culture (Table 1). Use of solution-phase hybridization has allowed tests to be performed singly or in batches in a familiar microwell format.
Although direct detection of organisms in clinical specimens by nucleic acid probes is rapid and simple, it suffers from lack of sensitivity. Most direct probe detection assays require at least 104 copies of nucleic acid per microliter for reliable detection, a requirement rarely met in clinical samples without some form of amplification. Amplification of the detection signal after probe hybridization improves sensitivity to as low as 500 gene copies per microliter and provides quantitative capabilities. This approach has been used extensively for quantitative assays of viral load (HIV, hepatitis B virus [HBV] and hepatitis C virus [HCV]) (Table 1) but does not match the analytical sensitivity of target amplification-based methods, such as polymerase chain reaction (PCR), for detecting organisms.
The commercial probe systems that use solution-phase hybridization and chemiluminescence for direct detection of infectious agents in clinical material include the PACE2 products of Gen-Probe and the hybrid capture assay systems of Digene and Murex (Table 1). These systems are user friendly, have a long shelf life, and are adaptable to small or large numbers of specimens. The PACE2 products are designed for direct detection of both Neisseria gonorrhoeae and Chlamydia trachomatis in a single specimen (one specimen, two separate probes). The hybrid capture systems detect human papilloma virus (HPV) in cervical scrapings, herpes simplex virus (HSV) in vesicle material, and cytomegalovirus (CMV) in blood and other fluids. All these tests have demonstrated sensitivity exceeding that of culture or immunologic methods for detecting the respective pathogens but are less sensitive than PCR or other target amplification-based methods.
The signal amplification-based probe methods for detection and quantitation of viruses (HBV, HCV, HIV) are presented in an enzyme immunoassay-like format and include branched chain DNA probes (Chiron) and QB replicase (Gene-Trak) methods (Table 1). These methods are not as sensitive as target amplification-based methods for detection of viruses; however, the quantitative results have proven useful for determining viral load and prognosis and for monitoring response to therapy (13).
Probe hybridization is useful for identifying slow-growing organisms after isolation in culture using either liquid or solid media. Identification of mycobacteria and other slow-growing organisms such as the dimorphic fungi (Histoplasma capsulatum, Coccidioides immitis, and Blastomyces dermatitidis) has certainly been facilitated by commercially available probes. All commercial probes for identifying organisms are produced by Gen-Probe and use acridinium ester-labeled probes directed at species-specific rRNA sequences (Table 1). Gen-Probe products are available for the culture identification of Mycobacterium tuberculosis, M. avium-intracellulare complex, M. gordonae, M. kansasii, Cryptococcus neoformans, the dimorphic fungi (listed above), N. gonorrhoeae, Staphylococcus aureus, Streptococcus pneumoniae, Escherichia coli, Haemophilus influenzae, Enterococcus spp., S. agalactiae, and Listeria monocytogenes. The sensitivity and specificity of these probes are excellent, and they provide species identification within one working day. Because most of the bacteria listed, plus C. neoformans, can be easily and efficiently identified by conventional methods within 1 to 2 days, many of these probes have not been widely used. The mycobacterial probes, on the other hand, are accepted as mainstays for the identification of M. tuberculosis and related species (7).
Nucleic acid amplification provides the ability to selectively amplify specific targets present in low concentrations to detectable levels; thus, amplification-based methods offer superior performance, in terms of sensitivity, over the direct (non-amplified) probe-based tests. PCR (Roche Molecular Systems, Branchburg, NJ) was the first such technique to be developed and because of its flexibility and ease of performance remains the most widely used molecular diagnostic technique in both research and clinical laboratories. Several different amplification-based strategies have been developed and are available commercially (Table 1). Commercial amplification-based molecular diagnostic systems for infectious diseases have focused largely on systems for detecting N. gonorrhoeae, C. trachomatis, M. tuberculosis, and specific viral infections (HBV, HCV, HIV, CMV, and enterovirus) (Table 1). Given the adaptability of PCR, numerous additional infectious pathogens have been detected by investigator-developed or home-brew PCR assays (5) (Table 2). In many instances, such tests provide important and clinically relevant information that would otherwise be unavailable since commercial interests have been slow to expand the line of products available to clinical laboratories. In addition to qualitative detection of viruses, quantitation of viral load in clinical specimens is now recognized to be of great importance for the diagnosis, prognosis, and therapeutic monitoring for HCV, HIV, HBV, and CMV (13). Both PCR and nucleic acid strand-based amplification systems are available for quantitation of one or more viruses (Table 1).
The adaptation of amplification-based test methods to commercially available kits has served to optimize user acceptability, prevent contamination, standardize reagents and testing conditions, and make automation a possibility. It is not clear to what extent the levels of detection achievable by the different amplification strategies differ. None of the newer methods provides a level of sensitivity greater than that of PCR. In choosing a molecular diagnostic system, one should consider the range of tests available, suitability of the method to workflow, and cost (6). Choosing one amplification-based method that provides testing capabilities for several pathogens is certainly practical.
Amplification-based methods are also valuable for identifying cultured and non-cultivatable organisms (5). Amplification reactions may be designed to rapidly identify an acid-fast organism as M. tuberculosis or may amplify a genus-specific or "universal" target, which then is characterized by using restriction endonuclease digestion, hybridization with multiple probes, or sequence determination to provide species or even subspecies delineation (4,5,14). Although identification was initially applied to slow-growing mycobacteria, it has applications for other pathogens that are difficult or impossible to identify with conventional methods.
Molecular methods can rapidly detect antimicrobial-drug resistance in clinical settings and have substantially contributed to our understanding of the spread and genetics of resistance (9). Conventional broth- and agar-based antimicrobial susceptibility testing methods provide a phenotypic profile of the response of a given microbe to an array of agents. Although useful for selecting potentially useful therapeutic agents, conventional methods are slow and fraught with problems. The most common failing is in the detection of methicillin resistance in staphylococci, which may be expressed in a very heterogeneous fashion, making phenotypic characterization of resistance difficult (9,15). Currently, molecular detection of the resistance gene, mec A, is the standard against which phenotypic methods for detection of methicillin resistance are judged (9,15,16).
Molecular methods may be used to detect specific antimicrobial-drug resistance genes (resistance genotyping) in many organisms (Table 3) (8,9). Detection of specific point mutations associated with resistance to antiviral agents is also increasingly important (17,18). Screening for mutations in an amplified product may be facilitated by the use of high-density probe arrays (Gene chips) (6).
Despite its many potential advantages, genotyping will not likely replace phenotypic methods for detecting antimicrobial-drug resistance in the clinical laboratory in the near future. Molecular methods for resistance detection may be applied directly to the clinical specimen, providing simultaneous detection and identification of the pathogen plus resistance characterization (9). Likewise, they are useful in detecting resistance in viruses, slow-growing or nonviable organisms, or organisms with resistance mechanisms that are not reliably detected by phenotypic methods (9,19). However, because of their high specificity, molecular methods will not detect newly emerging resistance mechanisms and are unlikely to be useful in detecting resistance genes in species where the gene has not been observed previously (19). Furthermore, the presence of a resistance gene does not mean that the gene will be expressed, and the absence of a known resistance gene does not exclude the possibility of resistance from another mechanism. Phenotypic antimicrobial susceptibility testing methods allow laboratories to test many organisms and detect newly emerging as well as established resistance patterns.
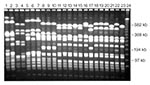
Laboratory characterization of microbial pathogens as biologically or genetically related is frequently useful in investigations (12,20,21). Several different epidemiologic typing methods have been applied in studies of microbial pathogens (Table 4). The phenotypic methods have occasionally been useful in describing the epidemiology of infectious diseases; however, they are too variable, slow, and labor-intensive to be of much use in most epidemiologic investigations. Newer DNA-based typing methods have eliminated most of these limitations and are now the preferred techniques for epidemiologic typing. The most widely used molecular typing methods include plasmid profiling, restriction endonuclease analysis of plasmid and genomic DNA, Southern hybridization analysis using specific DNA probes, and chromosomal DNA profiling using either pulsed-field gel electrophoresis (PFGE) or PCR-based methods (12,20). All these methods use electric fields to separate DNA fragments, whole chromosomes, or plasmids into unique patterns or fingerprints that are visualized by staining with ethidium bromide or by nucleic acid probe hybridization (Figure). Molecular typing is performed to determine whether different isolates give the same or different results for one or more tests. Epidemiologically related isolates share the same DNA profile or fingerprint, whereas sporadic or epidemiologically unrelated isolates have distinctly different patterns (Figure). If isolates from different patients share the same fingerprint, they probably originated from the same clone and were transmitted from patient to patient by a common source or mechanism.
Molecular typing methods have allowed investigators to study the relationship between colonizing and infecting isolates in individual patients, distinguish contaminating from infecting strains, document nosocomial transmission in hospitalized patients, evaluate reinfection versus relapse in patients being treated for an infection, and follow the spread of antimicrobial-drug resistant strains within and between hospitals over time (12). Most available DNA-based typing methods may be used in studying nosocomial infections when applied in the context of a careful epidemiologic investigation (12,21). In contrast, even the most powerful and sophisticated typing method, if used indiscriminately in the absence of sound epidemiologic data, may provide conflicting and confusing information.
Molecular testing for infectious diseases includes testing for the host's predisposition to disease, screening for infected or colonized persons, diagnosis of clinically important infections, and monitoring the course of infection or the spread of a specific pathogen in a given population. It is often assumed that in addition to improved patient care, major financial benefits may accrue from molecular testing because the tests reduce the use of less sensitive and specific tests, unnecessary diagnostic procedures and therapies, and nosocomial infections (11). However, the inherent costs of molecular testing methods, coupled with variable and inadequate reimbursement by third-party payers and managed-care organizations, have limited the introduction of these tests into the clinical diagnostic laboratory.
Not all molecular diagnostic tests are extremely expensive. Direct costs vary widely, depending on the test's complexity and sophistication. Inexpensive molecular tests are generally kit based and use methods that require little instrumentation or technologist experience. DNA probe methods that detect C. trachomatis or N. gonorrhoeae are examples of low-cost molecular tests. The more complex molecular tests, such as resistance genotyping, often have high labor costs because they require experienced, well-trained technologists. Although the more sophisticated tests may require expensive equipment (e.g., DNA sequencer) and reagents, advances in automation and the production of less-expensive reagents promise to decrease these costs as well as technician time. Major obstacles to establishing a molecular diagnostics laboratory that are often not considered until late in the process are required licenses, existing and pending patents, test selection, and billing and reimbursement (22).
Reimbursement issues are a major source of confusion, frustration, and inconsistency. Reimbursement by third-party payers is confounded by lack of Food and Drug Administration (FDA) approval and Current Procedural Terminology (CPT) codes for many molecular tests. In general, molecular tests for infectious diseases have been more readily accepted for reimbursement; however, reimbursement is often on a case-by-case basis and may be slow and cumbersome. FDA approval of a test improves the likelihood that it will be reimbursed but does not ensure that the amount reimbursed will equal the cost of performing the test.
Perhaps more than other laboratory tests, molecular tests may be negatively affected by fee-for-service managed-care contracts and across-the-board discounting of laboratory test fees. Such measures often result in reimbursement that is lower than the cost of providing the test. Although molecular tests may be considered a means of promoting patient wellness, the financial benefits of patient wellness are not easily realized in the short term (11). Health maintenance organizations (HMOs) and managed-care organizations often appear to be operating on shorter time frames, and their administrators may not be interested in the long-term impact of diagnostic testing strategies.
Molecular screening programs for infectious diseases are developed to detect symptomatic and asymptomatic disease in individuals and groups. Persons at high risk, such as immunocompromised patients or those attending family planning or obstetrical clinics, are screened for CMV and Chlamydia, respectively. Likewise, all blood donors are screened for bloodborne pathogens. The financial outcome of such testing is unknown. The cost must be balanced against the benefits of earlier diagnosis and treatment and societal issues such as disease epidemiology and population management.
One of the most highly touted benefits of molecular testing for infectious diseases is the promise of earlier detection of certain pathogens. The rapid detection of M. tuberculosis directly in clinical specimens by PCR or other amplification-based methods is quite likely to be cost-effective in the management of tuberculosis (7). Other examples of infectious disease that are amenable to molecular diagnosis and for which management can be improved by this technology include HSV encephalitis, Helicobacter pylori infection, and neuroborreliosis caused by Borrelia burgdorferi. For HSV encephalitis, detection of HSV in cerebrospinal fluid (CSF) can direct specific therapy and eliminate other tests including brain biopsy. Likewise, detection of H. pylori in gastric fluid can direct therapy and obviate the need for endoscopy and biopsy. PCR detection of B. burgdorferi in CSF is helpful in differentiating neuroborreliosis from other chronic neurologic conditions and chronic fatigue syndrome.
As discussed earlier, molecular tests may be used to predict disease response to specific antimicrobial therapy. Detection of specific resistance genes (mec A, van A) or point mutations resulting in resistance has proven efficacious in managing disease. Molecular-based viral load testing has become standard practice for patients with chronic hepatitis and AIDS. Viral load testing and genotyping of HCV are useful in determining the use of expensive therapy such as interferon and can be used to justify decisions on extent and duration of therapy. With AIDS, viral load determinations plus resistance genotyping have been used to select among the various protease inhibitor drugs available for treatment, improving patient response and decreasing incidence of opportunistic infections.
Pharmacogenomics is the use of molecular-based tests to predict the response to specific therapies and to monitor the response of the disease to the agents administered. The best examples of pharmacogenomics in infectious diseases are the use of viral load and resistance genotyping to select and monitor antiviral therapy of AIDS and chronic hepatitis (17,18). This application improves disease outcome; shortens length of hospital stay; reduces adverse events and toxicity; and facilitates cost-effective therapy by avoiding unnecessary expensive drugs, optimizing doses and timing, and eliminating ineffective drugs.
Molecular strain typing of microorganisms is now well recognized as an essential component of a comprehensive infection control program that also involves the infection control department, the infectious disease division, and pharmacy (10,21). Molecular techniques for establishing presence or absence of clonality are effective in tracking the spread of nosocomial infections and streamlining the activities of the infection control program (21,23). A comprehensive infection control program uses active surveillance by both infection control practitioners and the clinical microbiology laboratory to identify clusters of infections with a common microbial phenotype (same species and antimicrobial susceptibility profile). The isolates are then characterized in the laboratory by using one of a number of molecular typing methods (Table 4) to confirm or refute clonality. Based on available epidemiologic and molecular data, the hospital epidemiologist then develops an intervention strategy. Molecular typing can shorten or prevent an epidemic (23) and reduce the number and cost of nosocomial infections (Table 5) (10). Hacek et al. (10) analyzed the medical and economic benefits of an infection control program that included routine determination of microbial clonality and found that nosocomial infections were significantly decreased and more than $4 million was saved over a 2-year period (Table 5).
The true financial impact of molecular testing will only be realized when testing procedures are integrated into total disease assessment. More expensive testing procedures may be justified if they reduce the use of less-sensitive and less-specific tests and eliminate unnecessary diagnostic procedures and ineffective therapies.
Dr. Pfaller is professor and director of the Molecular Epidemiology and Fungus Testing Laboratory at the University of Iowa College of Medicine and College of Public Health. His research focuses on the epidemiology of nosocomial infections and antimicrobial-drug resistance.
References
- Cormican MG, Pfaller MA. Molecular pathology of infectious diseases. In: Henry JB, editor. Clinical diagnosis and management by laboratory methods. 19th ed. Philadelphia: W.B. Saunders Company; 1996:1390-9.
- Pfaller MA. Diagnosis and management of infectious diseases: Molecular methods for the new millennium. Clinical Laboratory News. 2000;26:10–3.
- Kant JA. Molecular diagnostics: Reimbursement and other selected financial issues. Diagn Mol Pathol. 1995;4:79–81. DOIPubMedGoogle Scholar
- Fredricks DN, Relman DA. Sequence-based identification of microbial pathogens: A reconsideration of Koch's postulates. Clin Microbiol Rev. 1996;9:18–33.PubMedGoogle Scholar
- Fredricks DN, Relman DA. Application of polymerase chain reaction to the diagnosis of infectious disease. Clin Infect Dis. 1999;29:475–88. DOIPubMedGoogle Scholar
- Tang YW, Persing DH. Molecular detection and identification of microorganisms. In: Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken RH, editors. Manual of clinical microbiology. 7th ed. Washington: American Society for Microbiology; 1999:215-44.
- Woods GL. Molecular techniques in mycobacterial detection. Arch Pathol Lab Med. 2001;125:122–6.PubMedGoogle Scholar
- Bergeron MG, Ouellette M. Preventing antibiotic resistance using rapid DNA-based diagnostic tests. Infect Control Hosp Epidemiol. 1998;19:560–4. DOIPubMedGoogle Scholar
- Cockerill FR III. Genetic methods for assessing antimicrobial resistance. Antimicrob Agents Chemother. 1999;43:199–212.PubMedGoogle Scholar
- Hacek DM, Suriano T, Noskin GA, Kruszynski J, Reisberg B, Peterson LR. Medical and economic benefit of a comprehensive infection control program that includes routine determination of microbial clonality. Am J Clin Pathol. 1999;111:647–54.PubMedGoogle Scholar
- Ross JS. Financial determinants of outcomes in molecular testing. Arch Pathol Lab Med. 1999;123:1071–5.PubMedGoogle Scholar
- Pfaller MA. Molecular epidemiology in the care of patients. Arch Pathol Lab Med. 1999;123:1007–10.PubMedGoogle Scholar
- Nolte FS. Impact of viral load testing on patient care. Arch Pathol Lab Med. 1999;123:1011–4.PubMedGoogle Scholar
- Anthony RM, Brown TJ, French GL. Rapid diagnosis of bacteremia by universal amplification of 23S ribosomal DNA followed by hybridization to an oligonucleotide array. J Clin Microbiol. 2000;38:781–8.PubMedGoogle Scholar
- Marshall SA, Wilke WW, Pfaller MA, Jones RN. Staphylococcus aureus and coagulase-negative staphylococci from blood stream infections: Frequency of occurrence, antimicrobial susceptibility, and molecular (mec A) characterization of oxacillin resistance in the SCOPE Program. Diagn Microbiol Infect Dis. 1998;30:205–14. DOIPubMedGoogle Scholar
- Hussain Z, Stoakes L, Massey V, Diagre D, Fitzgerald V, El Sayed S, Correlation of oxacillin MIC with mec A gene carriage in coagulase-negative staphylococci. J Clin Microbiol. 2000;38:752–4.PubMedGoogle Scholar
- Hecht FM, Grant RM, Petropoulos CJ, Dillon B, Chesney MA, Tian H, Sexual transmission of an HIV-1 variant resistant to multiple reverse-transcriptase and protease inhibitors. N Engl J Med. 1998;339:307–11. DOIPubMedGoogle Scholar
- Stuyver L, Van Geyt C, de Gendt S, Van Reybroeck G, Zoulin F, Leroux-Rods G, Line probe assay for monitoring drug resistance in hepatitis B virus-infected patients during antiviral therapy. J Clin Microbiol. 2000;38:702–7.PubMedGoogle Scholar
- Courvalin P. Genotypic approach to the study of bacterial resistance to antibiotics. Antimicrob Agents Chemother. 1991;35:1019–23.PubMedGoogle Scholar
- Arbeit RD. Laboratory procedures for epidemiologic analysis of microorganisms. In: Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken RH, editors. Manual of clinical microbiology. 7th ed. Washington: American Society for Microbiology; 1999:116-37.
- Pfaller MA, Herwaldt LA. The clinical microbiology laboratory and infection control: Emerging pathogens, antimicrobial resistance, and new technology. Clin Infect Dis. 1997;25:858–70. DOIPubMedGoogle Scholar
- Ferreira-Gonzalez A, Garrett CG. Pitfalls in establishing a molecular diagnostic laboratory. Hum Pathol. 1996;27:437–40. DOIPubMedGoogle Scholar
- Back NA, Linnemann CC, Pfaller MA, Staneck JL, Morthland V. Recurrent epidemics caused by a single strain of erythromycin-resistant Staphylococcus aureus: The importance of molecular epidemiology. JAMA. 1993;270:1329–33. DOIPubMedGoogle Scholar
Figure
Tables
Cite This ArticleTable of Contents – Volume 7, Number 2—April 2001
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Michael Pfaller, Medical Microbiology Division, C606 GH, Department of Pathology, University of Iowa College of Medicine, Iowa City, Iowa 52242, USA; fax:319-356-4916
Top