Volume 8, Number 11—November 2002
THEME ISSUE
Tuberculosis Genotyping
Tuberculosis Genotyping Network, United States
National Tuberculosis Genotyping and Surveillance Network: Analysis of the Genotype Database
Figure 7
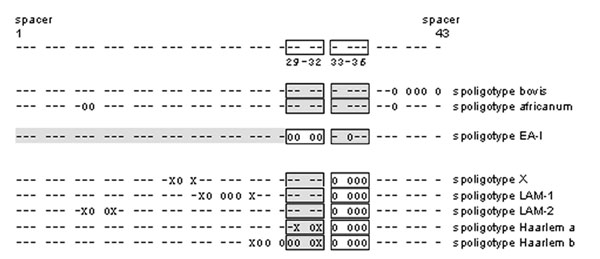
Figure 7. Motifs used to assign spoligotype patterns to spoligotype families. Each spoligotype was analyzed for the bovis (16), africanum (17), East-African-Indian (EA-I) (13,18), X (13), Latin American-Mediterranean 1 and 2 (13,18), and Haarlem a and b spoligotype motifs (10). Each motif definition was modified from the original references to ensure that motifs were not identified in a spoligotype pattern due to an unrelated deletion at the spacers of interest; each of the motif-defining absent spacers must be flanked on both sides by the adjacent spacer. The 43 spacers in the spoligotype pattern are classified with symbols: X: spacer must be present; 0: spacer must be absent; -: spacer may or may not be present; spacers in shaded boxes: at least one of the spacers in the box must be present.
References
- Crawford JT, Braden CR, Schable BA, Onorato IM. National Tuberculosis Genotyping and Surveillance Network: design and methods. Emerg Infect Dis. 2002;8:1192–6.PubMedGoogle Scholar
- Dale JW, Brittain D, Cataldi AA, Cousins D, Crawford JT, Driscoll J, Spacer oligonucleotide typing of bacteria of the Mycobacterium tuberculosis complex: recommendations for standardized nomenclature. Int J Tuberc Lung Dis. 2001;5:216–9.PubMedGoogle Scholar
- Yang Z, Barnes PF, Chaves F, Eisenach KD, Weis SE, Bates JH, Diversity of DNA fingerprints of Mycobacterium tuberculosis isolates in the United States. J Clin Microbiol. 1998;36:1003–7.PubMedGoogle Scholar
- Bifani PJ, Mathema B, Kurepina NE, Kreiswirth BN. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 2002;10:45–52. DOIPubMedGoogle Scholar
- van Soolingen D, Qian L, de Haas PE, Douglas JT, Traore H, Portaels F, Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J Clin Microbiol. 1995;33:3234–8.PubMedGoogle Scholar
- Beggs ML, Eisenach KD, Cave MD. Mapping of IS6110 insertion sites in two epidemic strains of Mycobacterium tuberculosis. J Clin Microbiol. 2000;38:2923–8.PubMedGoogle Scholar
- Steinlein LM, Crawford JT. Reverse dot blot assay (insertion site typing) for precise detection of sites of IS6110 insertion in the Mycobacterium tuberculosis genome. J Clin Microbiol. 2001;39:871–8. DOIPubMedGoogle Scholar
- Bifani P, Moghazeh S, Shopsin B, Driscoll J, Ravikovitch A, Kreiswirth BN. Molecular characterization of Mycobacterium tuberculosis H37Rv/Ra variants: distinguishing the mycobacterial laboratory strain. J Clin Microbiol. 2000;38:3200–4.PubMedGoogle Scholar
- Kremer K, van Soolingen D, Frothingham R, Haas WH, Hermans PWM, Martin C, Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J Clin Microbiol. 1999;37:2607–18.PubMedGoogle Scholar
- Cowan LS, Mosher L, Diem L, Massey JP, Crawford JT. Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low-copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J Clin Microbiol. 2002;40:1592–602. DOIPubMedGoogle Scholar
- Plikaytis BB, Kurepina N, Woodley CL, Butler WR, Shinnick TM. Multiplex PCR assay to aid in the identification of the highly transmissible Mycobacterium tuberculosis strain CDC1551. Tuber Lung Dis. 1999;79:273–8. DOIPubMedGoogle Scholar
- Sebban M, Mokrousov I, Rastogi N, Sola C. A data-mining approach to spacer oligonucleotide typing of Mycobacterium tuberculosis. Bioinformatics. 2002;18:235–43. DOIPubMedGoogle Scholar
- Soini H, Pan X, Amin A, Graviss EA, Siddiqui A, Musser JM. Characterization of Mycobacterium tuberculosis isolates from patients in Houston, Texas, by spoligotyping. J Clin Microbiol. 2000;38:669–76.PubMedGoogle Scholar
- Sreevatsan S, Pan X, Stockbauer K, Connell ND, Kreiswirth BN, Whittam TS, Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci U S A. 1997;94:9869–74. DOIPubMedGoogle Scholar
- Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–14.PubMedGoogle Scholar
- Viana-Niero C, Gutierrez C, Sola C, Filliol I, Boulahbal F, Vincent V, Genetic diversity of Mycobacterium africanum clinical isolates based on IS6110-restriction fragment length polymorphism analysis, spoligotyping, and variable number of tandem DNA repeats. J Clin Microbiol. 2001;39:57–65. DOIPubMedGoogle Scholar
- Sola C, Filliol I, Legrand E, Mokrousov I, Rastogi N. Mycobacterium tuberculosis phylogeny reconstruction based on combined numerical analysis with IS1081, IS6110, VNTR, and DR-based spoligotyping suggests the existence of two new phylogeographical clades. J Mol Evol. 2001;53:680–9. DOIPubMedGoogle Scholar
- Mathema B, Bifani PJ, Driscoll J, Steinlein L, Kurepina N, Moghazeh SL, Identification and evolution of an IS6110 low-copy Mycobacterium tuberculosis cluster. J Infect Dis. 2002;185:641–9. DOIPubMedGoogle Scholar
- Fomukong N, Beggs M, El Hajj H, Templeton G, Eisenach K, Cave MD. Differences in the prevalence of IS6110 insertion sites in Mycobacterium tuberculosis strains: low and high-copy numbers of IS6110. Tuber Lung Dis. 1998;78:109–16. DOIGoogle Scholar