Volume 14, Number 11—November 2008
Dispatch
Metagenomic Diagnosis of Bacterial Infections
Figure
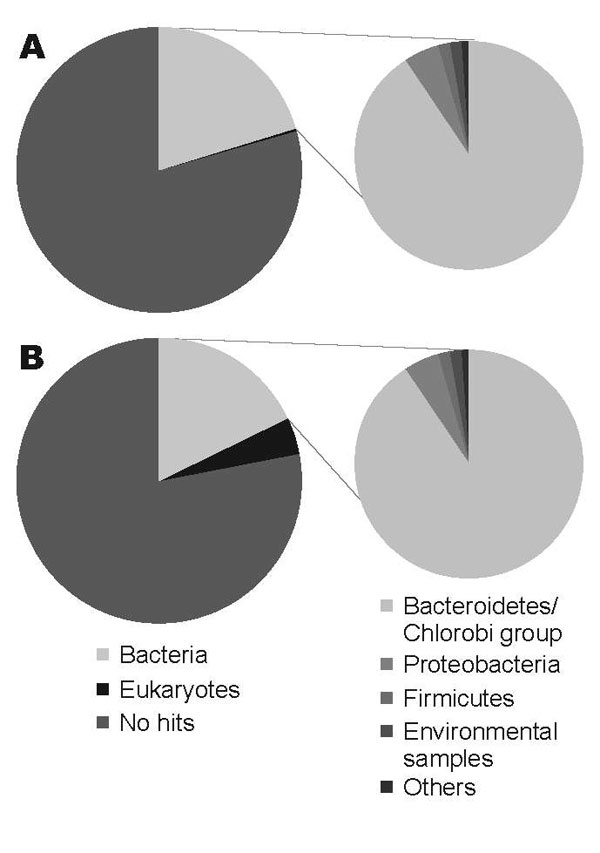
Figure. Comparison of the organisms from which the best matches for the sequences were derived from a BLASTN (http://blast.ncbi.nlm.nih.gov) search with an expect-value cutoff of 10–5. A) DNA from nondiarrheic fecal sample collected 3 months after patient had recovered. B) DNA from diarrheic fecal sample collected while patient was ill.
Page created: July 21, 2010
Page updated: July 21, 2010
Page reviewed: July 21, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.