Volume 14, Number 4—April 2008
Research
Hemorrhagic Fever with Renal Syndrome Caused by 2 Lineages of Dobrava Hantavirus, Russia1
Figure 2
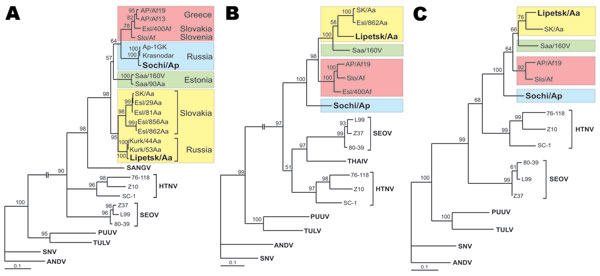
Figure 2. Maximum-likelihood phylogenetic trees of Dobrava-Belgrade virus (DOBV), showing the phylogenetic placement of novel Russian isolates Sochi/Ap and Lipetsk/Aa (in boldface) based on A) complete S-segment open reading frame (ORF) (nucleotide sequence positions 36–1325), B) complete M-segment ORF (nt positions 41–3445), and C) partial L-segment sequences (374 nt, positions 157–530). Different DOBV lineages are marked by colored boxes: yellow, DOBV-Aa; green, Saaremaa; red, DOBV-Af; blue, DOBV-Ap (Sochi virus). The Sochi/Ap and Lipetsk/Aa S-, M-, and L-segment sequences were deposited in GenBank under accession nos. EU188449–EU188454. The trees were computed with the TREE-PUZZLE package by using the Tamura Nei evolutionary model. The values at the tree branches are the PUZZLE support values. The scale bar indicates an evolutionary distance of 0.1 substitutions per position in the sequence. SANGV, Sangassou virus; HTNV, Hantaan virus; SEOV, Seoul virus; PUUV, Puumala virus; TULV, Tula virus; SNV, Sin Nombre virus, ANDV, Andes virus; THAIV, Thailand hantavirus.
1This work is dedicated to the memory of our friend and colleague Milan Labuda, who died in August 2007.