Volume 14, Number 9—September 2008
Dispatch
Juquitiba-like Hantavirus from 2 Nonrelated Rodent Species, Uruguay
Figure 1
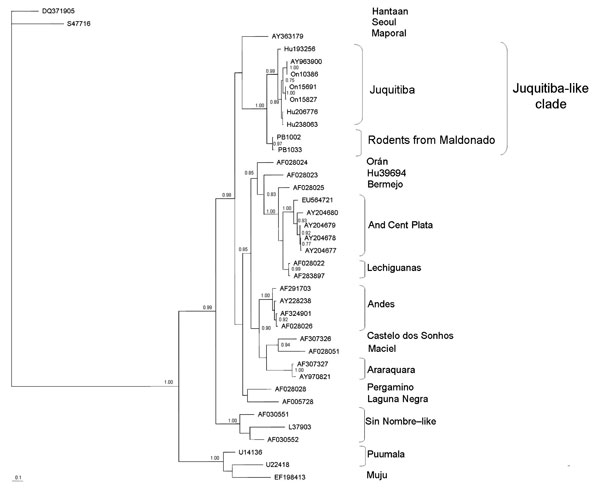
Figure 1. Majority-rule consensus tree obtained in the Bayesian analysis of sequences of the medium segment of Juquitiba-like hantavirus isolated from 2 nonrelated rodent species. Posterior probabilities >0.80 are shown at the nodes. Alignment and editing of nucleotide sequences were conducted by using BioEdit v7.0.9.0 (www.mbio.ncsu.edu/BioEdit/BioEdit.html). Sequences of Seoul and Hantaan hantaviruses were used as outgroup. Estimation of the suitable model of nucleotide substitution and phylogenetic analyses were carried out by using Modelgenerator (http://bioinf.may.ie/software/modelgenerator), MrBayes v3.1.2 (Bayesian analysis; http://mrbayes.csit.fsu.edu), and PAUP* 4.0b10 (maximum-parsimony analysis; http://paup.csit.fsu.edu). Bayesian analyses were conducted under the general time reversible + gamma + proportion invariant model. Two runs of 4 chains each (1 cold, 3 heated, temperature 0.20) were run for 3 million generations; trees were sampled every 100 generations. Convergence was assessed by using the average standard deviation in partition frequency values across independent analyses with a threshold value of 0.01; burn-in was set to 25%. Seropositive specimens from Uruguay are as follows: PB1033 (black-footed pigmy rice rat, Oligoryzomys nigripes) and PB1002 (long-nosed mouse, Oxymycterus nasutus), GenBank accession nos. EU564726 and EU564725, respectively. Analyzed hantavirus sequences are Hantaan, (DQ371905), Seoul (SA7716), Juquitiba (AY963900, On10386, On15691, On15827, Hu206776, Hu238063, Hu193256), Maporal (AY363179), Andes Central Plata (AY204678, AY204677, AY204679, AY204680, EU564721), Lechiguanas (AF028022, AF283897), Bermejo (AF028025), Hu39694 (AF028023), Orán (AF028024), Andes (AF291703, AF324901, AF028026, AY228238), Castelo dos Sonhos (AF307326), Maciel (AF028051), Araraquara (AF307327, AY970821), Pergamino (AF028028), Laguna Negra (AF005728), Sin Nombre–like (L37903, AF030552, AF030551), Puumala (U14136, U22418), and Muju (EF198413). Scale bar indicates expected changes per site.