Volume 15, Number 12—December 2009
Dispatch
Bartonella rochalimae in Raccoons, Coyotes, and Red Foxes
Figure 2
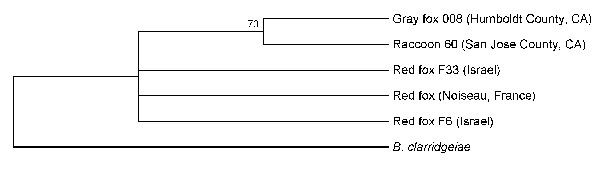
Figure 2. Phylogenetic tree of Bartonella species based on intergenic transcribed spacer sequence alignment for the isolates from the gray foxes, red foxes, and raccoons. Raccoon and gray fox isolates are shown for comparison. The tree shown is a neighbor-joining tree based on the Kimura 2-parameter model of nucleotide substitution. Bootstrap values are based on 1,000 replicates. The analysis provided tree topology only; the lengths of the vertical and horizontal lines are not significant.
Page created: December 09, 2010
Page updated: December 09, 2010
Page reviewed: December 09, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.