Volume 15, Number 3—March 2009
Research
Shiga Toxin–producing Escherichia coli Strains Negative for Locus of Enterocyte Effacement
Figure 1
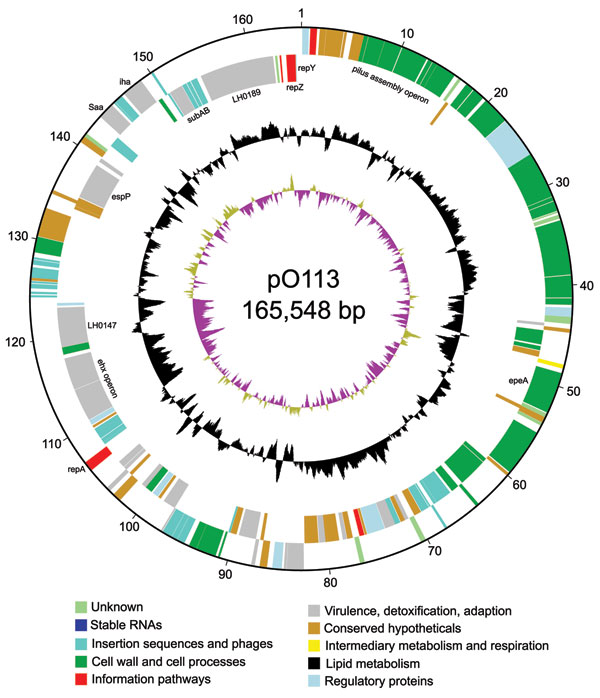
Figure 1. Circular map of virulence plasmid pO113 generated by using circular_diagram.pl (Sanger Institute, Cambridge, UK) and Inkscape software (www.inkscape.org). The locations of proteins encoded on the leading and lagging strand are shown on the outer 2 rings. The colors indicate the assigned GenoList functional category (Institut Pasteur, Paris, France). The black ring indicates GC content, with high GC content outermost. The innermost ring shows GC skew.
Page created: December 07, 2010
Page updated: December 07, 2010
Page reviewed: December 07, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.