Volume 15, Number 3—March 2009
Research
Shiga Toxin–producing Escherichia coli Strains Negative for Locus of Enterocyte Effacement
Figure 4
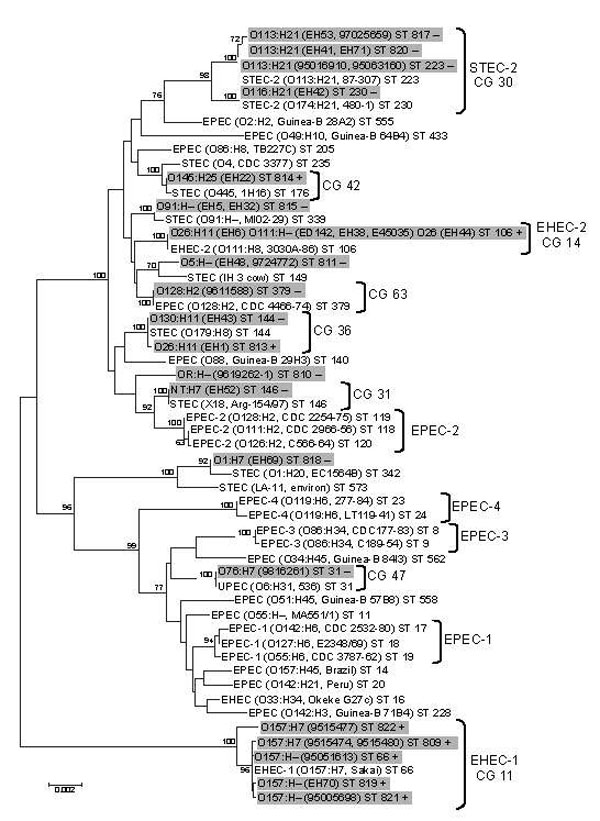
Figure 4. Phylogenetic relationships of 17 locus of enterocyte effacement (LEE)–negative and 13 LEE-positive Shiga toxin–producing Escherichia coli (STEC) strains (highlighted in gray) compared with a cohort of reference E. strains. Phylogeny was demonstrated by a neighbor-joining algorithm from 7 housekeeping gene sequences. Each isolate has been assigned a sequence type (ST) (in boldface), and assigned clonal groups (CGs) are displayed. The scale bar demonstrates the branch length that corresponds to 2 nucleotide substitutions per 1,000 nucleotide sites. Significant nodes were identified by bootstrapping (Monte Carlo randomization); nodes were present in >70% of the 1,000 bootstrap trees highlighted and identified as significant.
Page created: December 07, 2010
Page updated: December 07, 2010
Page reviewed: December 07, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.