Volume 16, Number 8—August 2010
Perspective
Hantavirus Infections in Humans and Animals, China
Figure 3
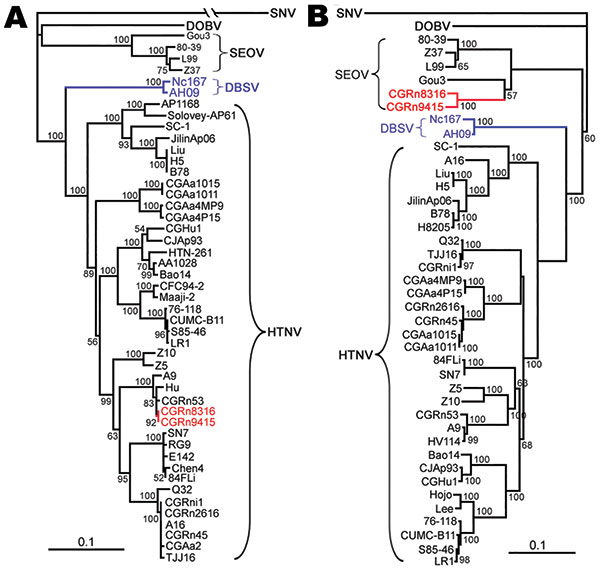
Figure 3. Phylogenetic trees of Hantaan virus (HTNV) variants according to the small segment (A) and medium segment (B) coding sequences. PHYLIP program package version 3.65 (http://helix.nih.gov/Applications/phylip.html) was used to construct the phylogenetic trees; the neighbor-joining method was used. Bootstrap values were calculated from 1,000 replicates; only values >50% are shown at the branch nodes. The trees constructed using the maximum-likelihood method (not shown) had similar topology. Scale bars indicate nucleotide substitutions per site. Colors (blue and red) highlight viruses of interest from China. SNV, Sin Nombre virus; DOBV, Dobrava-Belgrade virus; SEOV, Seoul virus; DBSV, Da Bie Shan virus.
Page created: March 30, 2011
Page updated: March 30, 2011
Page reviewed: March 30, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.