Volume 5, Number 1—February 1999
Research
Dual and Recombinant Infections: An Integral Part of the HIV-1 Epidemic in Brazil
Figure 2
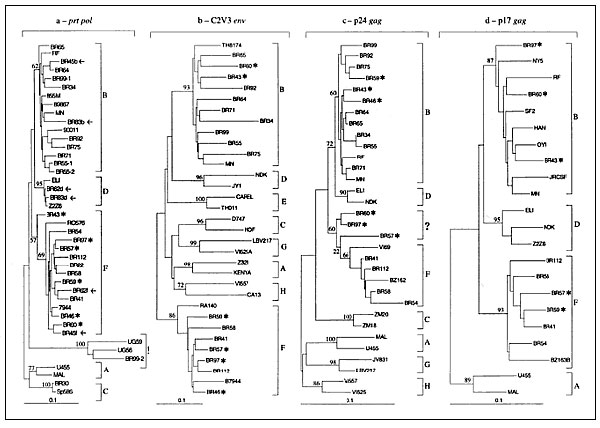
Figure 2. Phylogenetic classification of HIV-1 sequences from Brazilian patients (denoted with BR prefix). The trees were constructed on the basis of DNA sequences of prt (a), env (b), gag-p24 (c), and gag-p17 (d) by the neighbor-joining method. Numbers at the branch nodes connected with subtypes indicate bootstrap values. An arrow indicates dually infected specimens; an asterisk shows viral sequences, which clustered into different lineages depending on which parts of viral genome were analyzed; ! represents hypermutated sequences; ? represents unclassified subtype of p24 gag sequences. The distinct HIV-1 subtypes are delineated. The scale bar indicates an evolutionary distance of 0.10 nucleotides per position in the sequence. Vertical distances are for clarity only. GenBank accession numbers: prt [AF099155-99171;AF079986-79989; AF079991; and AF079994-79996]; env [AF113560-113576]; p17gag [AF115443-115451]; p24gag [AF115780-115797].