Volume 8, Number 6—June 2002
Dispatch
Three Drinking-Water–Associated Cryptosporidiosis Outbreaks, Northern Ireland
Figure
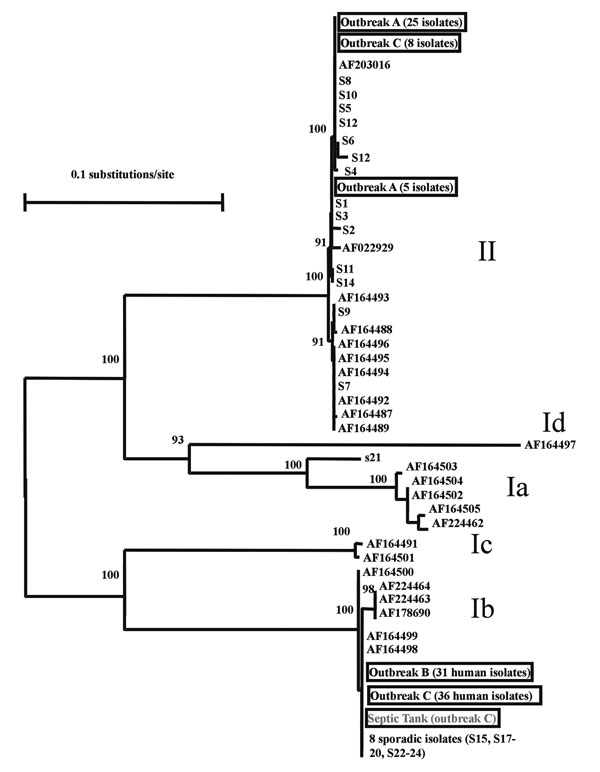
Figure. Genetic relationship among Cryptosporidium parasites found in three Northern Ireland outbreaks (outbreaks A, B, and C), sporadic cases in west Ireland (S1 to S14) and the northwest of England (S15 to S24), subgenotypes described by Strong et al. (11), and an unpublished sequence (AF203016) from the GenBank database. The isolates with accession numbers were mostly human and bovine and from the United States with the exception of AF164488, AF164492, and AF164493, which were isolated from humans in Zaire, Peru, and Brazil, respectively, but had been passaged in calves in the United States. Nomenclature for groups of subgenotypes is adapted from Strong et al.: Ia, Ib, Ic, and Id for subgenotypes of the C. parvum human genotype and II for subgenotypes of the C. parvum bovine genotype (11). Data presented are a neighbor-joining tree of GP60 sequences.
References
- Peng MM, Xiao L, Freeman AR, Arrowood MJ, Escalante AA, Weltman AC, Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg Infect Dis. 1997;3:567–73.PubMedGoogle Scholar
- McLauchlin J, Amar C, Pedraza-Diaz S, Nichols GL. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Cryptosporidium spp. in 1,705 fecal samples from humans and 105 fecal samples from livestock animals. J Clin Microbiol. 2000;38:3984–90.PubMedGoogle Scholar
- Sulaiman IM, Lal AA, Xiao L. A population genetic study of the Cryptosporidium parvum human genotype parasites. J Eukaryot Microbiol. 2001. In press.
- Peng MM, Matos O, Gatei W, Das P, Stantic-Pavlinic M, Bern C, A comparison of Cryptosporidium subgenotypes from several geographic regions. J Eukaryot Microbiol. 2001. In press.
- Sulaiman I, Xiao L, Yang C, Escalante L, Moore A, Beard CB, Differentiating human from animal isolates of Cryptosporidium parvum. Emerg Infect Dis. 1998;4:681–5.PubMedGoogle Scholar
- Ong CSL, Eisler DL, Goh SH, Tomblin J, Awad-El-Kariem FM, Beard CB, Molecular epidemiology of cryptosporidiosis outbreaks and transmission in British Columbia, Canada. Am J Trop Med Hyg. 1999;61:63–9.PubMedGoogle Scholar
- Communicable Disease Surveillance Centre. Foodborne and other gastro-intestinal outbreaks reported to CDSC (NI), Jan–June 2000. Communicable Diseases Monthly Report Northern Ireland Edition 2000;9:1.
- Communicable Disease Surveillance Centre. Cryptosporidiosis incident. Communicable Diseases Monthly Report Northern Ireland Edition 2000;9:1.
- Communicable Disease Surveillance Centre. Outbreak of cryptosporidiosis. Communicable Diseases Monthly Report Northern Ireland Edition 2001;10:1–2.
- Spano F, Putignani L, McLauchlin J, Casemore DP, Crisanti A. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol Lett. 1997;150:209–17.PubMedGoogle Scholar
- Ryley JF, Meade R, Hazelhurst J, Robinson TE. Methods in coccidiosis research: separation of oocysts from faeces. Parasitology. 1976;73:311. DOIPubMedGoogle Scholar
- Xiao L, Bern C, Limor J, Sulaiman I, Roberts J, Checkley W, Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J Infect Dis. 2001;183:492–7. DOIPubMedGoogle Scholar
- Strong WB, Gut J, Nelson RG. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect Immun. 2000;68:4117–34. DOIPubMedGoogle Scholar
- Xiao L, Singh A, Limor J, Graczyk TK, Gradus S, Lal AA. Molecular characterization of Cryptosporidium oocysts in samples of raw surface and wastewater. Appl Environ Microbiol. 2001;67:1097–101. DOIPubMedGoogle Scholar
- Xiao L, Escalante L, Yang C, Sulaiman IM, Escalante AA, Montali R, Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit RNA gene locus. Appl Environ Microbiol. 1999;65:1578–83.PubMedGoogle Scholar
- Xiao L, Alderisio K, Limor J, Royer M, Lal AA. Identification of species and sources of Cryptosporidium oocysts in storm waters with a small subunit rRNA-based diagnostic and genotyping tool. Appl Environ Microbiol. 2000;66:5492–8. DOIPubMedGoogle Scholar